Viral Ubiquitin Ligase Stimulates Selective Host MicroRNA Expression by Targeting ZEB Transcriptional Repressors
- PMID: 28783105
- PMCID: PMC5580467
- DOI: 10.3390/v9080210
Viral Ubiquitin Ligase Stimulates Selective Host MicroRNA Expression by Targeting ZEB Transcriptional Repressors
Abstract
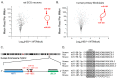
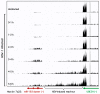
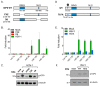
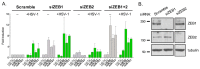
Infection with herpes simplex virus-1 (HSV-1) brings numerous changes in cellular gene expression. Levels of most host mRNAs are reduced, limiting synthesis of host proteins, especially those involved in antiviral defenses. The impact of HSV-1 on host microRNAs (miRNAs), an extensive network of short non-coding RNAs that regulate mRNA stability/translation, remains largely unexplored. Here we show that transcription of the miR-183 cluster (miR-183, miR-96, and miR-182) is selectively induced by HSV-1 during productive infection of primary fibroblasts and neurons. ICP0, a viral E3 ubiquitin ligase expressed as an immediate-early protein, is both necessary and sufficient for this induction. Nuclear exclusion of ICP0 or removal of the RING (really interesting new gene) finger domain that is required for E3 ligase activity prevents induction. ICP0 promotes the degradation of numerous host proteins and for the most part, the downstream consequences are unknown. Induction of the miR-183 cluster can be mimicked by depletion of host transcriptional repressors zinc finger E-box binding homeobox 1 (ZEB1)/-crystallin enhancer binding factor 1 (δEF1) and zinc finger E-box binding homeobox 2 (ZEB2)/Smad-interacting protein 1 (SIP1), which we establish as new substrates for ICP0-mediated degradation. Thus, HSV-1 selectively stimulates expression of the miR-183 cluster by ICP0-mediated degradation of ZEB transcriptional repressors.
Keywords: E3 ubiquitin ligase; HSV-1; ICP0; ZEB; herpes simplex virus; host shutoff; miR-182; miR-183; miR-96; microRNA.
Conflict of interest statement
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





Similar articles
-
A Tale of Two PMLs: Elements Regulating a Differential Substrate Recognition by the ICP0 E3 Ubiquitin Ligase of Herpes Simplex Virus 1.J Virol. 2016 Nov 14;90(23):10875-10885. doi: 10.1128/JVI.01636-16. Print 2016 Dec 1. J Virol. 2016. PMID: 27681131 Free PMC article.
-
Characterization of Elements Regulating the Nuclear-to-Cytoplasmic Translocation of ICP0 in Late Herpes Simplex Virus 1 Infection.J Virol. 2018 Jan 2;92(2):e01673-17. doi: 10.1128/JVI.01673-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29093084 Free PMC article.
-
Discovery of Small-Molecule Inhibitors Targeting the E3 Ubiquitin Ligase Activity of the Herpes Simplex Virus 1 ICP0 Protein Using an In Vitro High-Throughput Screening Assay.J Virol. 2019 Jun 14;93(13):e00619-19. doi: 10.1128/JVI.00619-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996104 Free PMC article.
-
The HSV-1 ubiquitin ligase ICP0: Modifying the cellular proteome to promote infection.Virus Res. 2020 Aug;285:198015. doi: 10.1016/j.virusres.2020.198015. Epub 2020 May 13. Virus Res. 2020. PMID: 32416261 Free PMC article. Review.
-
Role of ND10 nuclear bodies in the chromatin repression of HSV-1.Virol J. 2016 Apr 5;13:62. doi: 10.1186/s12985-016-0516-4. Virol J. 2016. PMID: 27048561 Free PMC article. Review.
Cited by
-
Genomic associations with poxvirus across divergent island populations in Berthelot's pipit.Mol Ecol. 2022 Jun;31(11):3154-3173. doi: 10.1111/mec.16461. Epub 2022 Apr 18. Mol Ecol. 2022. PMID: 35395699 Free PMC article.
-
Herpes Simplex Virus 1 Deregulation of Host MicroRNAs.Noncoding RNA. 2018 Nov 23;4(4):36. doi: 10.3390/ncrna4040036. Noncoding RNA. 2018. PMID: 30477082 Free PMC article. Review.
-
HSV-1 single-cell analysis reveals the activation of anti-viral and developmental programs in distinct sub-populations.Elife. 2019 May 15;8:e46339. doi: 10.7554/eLife.46339. Elife. 2019. PMID: 31090537 Free PMC article.
-
Neuronal miR-9 promotes HSV-1 epigenetic silencing and latency by repressing Oct-1 and Onecut family genes.Nat Commun. 2024 Mar 5;15(1):1991. doi: 10.1038/s41467-024-46057-6. Nat Commun. 2024. PMID: 38443365 Free PMC article.
-
Impact of HVT Vaccination on Splenic miRNA Expression in Marek's Disease Virus Infections.Genes (Basel). 2019 Feb 5;10(2):115. doi: 10.3390/genes10020115. Genes (Basel). 2019. PMID: 30764490 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007647/AI/NIAID NIH HHS/United States
- F31 DC011687/DC/NIDCD NIH HHS/United States
- R01 NS082240/NS/NINDS NIH HHS/United States
- T32 GM007308/GM/NIGMS NIH HHS/United States
- R01 DC002756/DC/NIDCD NIH HHS/United States
- R21 AI103933/AI/NIAID NIH HHS/United States
- R21 AI105896/AI/NIAID NIH HHS/United States
- P01 AI098681/AI/NIAID NIH HHS/United States
- P01 NS035138/NS/NINDS NIH HHS/United States
- T32 AI007180/AI/NIAID NIH HHS/United States
- R01 AI026126/AI/NIAID NIH HHS/United States
- P30 CA023168/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

