Potent and broad HIV-neutralizing antibodies in memory B cells and plasma
- PMID: 28783671
- PMCID: PMC5905719
- DOI: 10.1126/sciimmunol.aal2200
Potent and broad HIV-neutralizing antibodies in memory B cells and plasma
Abstract
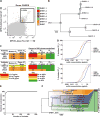
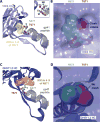
Induction of broadly neutralizing antibodies (bnAbs) is a goal of HIV-1 vaccine development. Antibody 10E8, reactive with the distal portion of the membrane-proximal external region (MPER) of HIV-1 gp41, is broadly neutralizing. However, the ontogeny of distal MPER antibodies and the relationship of memory B cell to plasma bnAbs are poorly understood. HIV-1-specific memory B cell flow sorting and proteomic identification of anti-MPER plasma antibodies from an HIV-1-infected individual were used to isolate broadly neutralizing distal MPER bnAbs of the same B cell clonal lineage. Structural analysis demonstrated that antibodies from memory B cells and plasma recognized the envelope gp41 bnAb epitope in a distinct orientation compared with other distal MPER bnAbs. The unmutated common ancestor of this distal MPER bnAb was autoreactive, suggesting lineage immune tolerance control. Construction of chimeric antibodies of memory B cell and plasma antibodies yielded a bnAb that potently neutralized most HIV-1 strains.
Copyright © 2017, American Association for the Advancement of Science.
Figures




References
-
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. - PMC - PubMed
-
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PWHI. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 2001;75:10892–10905. - PMC - PubMed
-
- Kwon YD, Georgiev IS, Ofek G, Zhang B, Asokan M, Bailer RT, Bao A, Caruso W, Chen X, Choe M, Druz A, Ko S-Y, Louder MK, McKee K, O’Dell S, Pegu A, Rudicell R, Shi W, Wang K, Yang Y, Alger M, Bender MF, Carlton K, Cooper JW, Blinn J, Eudailey J, Lloyd K, Parks R, Alam SM, Haynes BF, Padte NN, Yu J, Ho DD, Huang J, Connors M, Schwartz RM, Mascola JR, Kwong PD. Optimization of the solubility of HIV-1-neutralizing antibody 10E8 through somatic variation and structure-based design. J. Virol. 2016;90:5899–5914. - PMC - PubMed
-
- Chen Y, Zhang J, Hwang K-K, Bouton-Verville H, Xia S-M, Newman A, Ouyang Y-B, Haynes BF, Verkoczy L. Common tolerance mechanisms, but distinct cross-reactivities associated with gp41 and lipids, limit production of HIV-1 broad neutralizing antibodies 2F5 and 4E10. J. Immunol. 2013;191:1260–1275. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

