Characterization of T and B cell repertoire diversity in patients with RAG deficiency
- PMID: 28783691
- PMCID: PMC5586490
- DOI: 10.1126/sciimmunol.aah6109
Characterization of T and B cell repertoire diversity in patients with RAG deficiency
Abstract
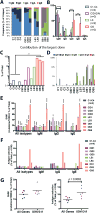
Recombination-activating genes 1 and 2 (RAG1 and RAG2) play a critical role in T and B cell development by initiating the recombination process that controls the expression of T cell receptor (TCR) and immunoglobulin genes. Mutations in the RAG1 and RAG2 genes in humans cause a broad spectrum of phenotypes, including severe combined immunodeficiency (SCID) with lack of T and B cells, Omenn syndrome, leaky SCID, and combined immunodeficiency with granulomas or autoimmunity (CID-G/AI). Using next-generation sequencing, we analyzed the TCR and B cell receptor (BCR) repertoire in 12 patients with RAG mutations presenting with Omenn syndrome (n = 5), leaky SCID (n = 3), or CID-G/AI (n = 4). Restriction of repertoire diversity skewed usage of variable (V), diversity (D), and joining (J) segment genes, and abnormalities of CDR3 length distribution were progressively more prominent in patients with a more severe phenotype. Skewed usage of V, D, and J segment genes was present also within unique sequences, indicating a primary restriction of repertoire. Patients with Omenn syndrome had a high proportion of class-switched immunoglobulin heavy chain transcripts and increased somatic hypermutation rate, suggesting in vivo activation of these B cells. These data provide a framework to better understand the phenotypic heterogeneity of RAG deficiency.
Copyright © 2016, American Association for the Advancement of Science.
Figures



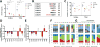

References
-
- Villa A, Santagata S, Bozzi F, Giliani S, Frattini A, Imberti L, Gatta LB, Ochs HD, Schwarz K, Notarangelo LD, Vezzoni P, Spanopoulou E. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. - PubMed
-
- Shearer WT, Dunn E, Notarangelo LD, Dvorak CC, Puck JM, Logan BR, Griffith LM, Kohn DB, O'Reilly RJ, Fleisher TA, Pai SY, Martinez CA, Buckley RH, Cowan MJ. Establishing diagnostic criteria for severe combined immunodeficiency disease (SCID), leaky SCID, and Omenn syndrome: the Primary Immune Deficiency Treatment Consortium experience. J Allergy Clin Immunol. 2014;133:1092–1098. - PMC - PubMed
-
- de Villartay JP, Lim A, Al-Mousa H, Dupont S, Dechanet-Merville J, Coumau-Gatbois E, Gougeon ML, Lemainque A, Eidenschenk C, Jouanguy E, Abel L, Casanova JL, Fischer A, Le Deist F. A novel immunodeficiency associated with hypomorphic RAG1 mutations and CMV infection. J Clin Invest. 2005;115:3291–3299. - PMC - PubMed
-
- Schuetz C, Huck K, Gudowius S, Megahed M, Feyen O, Hubner B, Schneider DT, Manfras B, Pannicke U, Willemze R, Knuchel R, Gobel U, Schulz A, Borkhardt A, Friedrich W, Schwarz K, Niehues T. An immunodeficiency disease with RAG mutations and granulomas. N Engl J Med. 2008;358:2030–2038. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

