SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana
- PMID: 28783755
- PMCID: PMC5544219
- DOI: 10.1371/journal.pone.0182591
SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana
Abstract
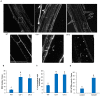
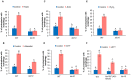
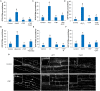
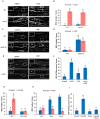
Autophagy is a degradation process in which cells break down and recycle their cytoplasmic contents when subjected to environmental stress or during cellular remodeling. The Arabidopsis thaliana SnRK1 complex is a protein kinase that senses changes in energy levels and triggers downstream responses to enable survival. Its mammalian ortholog, AMPK, and yeast ortholog, Snf-1, activate autophagy in response to low energy conditions. We therefore hypothesized that SnRK1 may play a role in the regulation of autophagy in response to nutrient or energy deficiency in Arabidopsis. To test this hypothesis, we determined the effect of overexpression or knockout of the SnRK1 catalytic subunit KIN10 on autophagy activation by abiotic stresses, including nutrient deficiency, salt, osmotic, oxidative, and ER stress. While wild-type plants had low basal autophagy activity in control conditions, KIN10 overexpression lines had increased autophagy under these conditions, indicating activation of autophagy by SnRK1. A kin10 mutant had a basal level of autophagy under control conditions similar to wild-type plants, but activation of autophagy by most abiotic stresses was blocked, indicating that SnRK1 is required for autophagy induction by a wide variety of stress conditions. In mammals, TOR is a negative regulator of autophagy, and AMPK acts to activate autophagy both upstream of TOR, by inhibiting its activity, and in a parallel pathway. Inhibition of Arabidopsis TOR leads to activation of autophagy; inhibition of SnRK1 did not block this activation. Furthermore, an increase in SnRK1 activity was unable to induce autophagy when TOR was also activated. These results demonstrate that SnRK1 acts upstream of TOR in the activation of autophagy in Arabidopsis.
Conflict of interest statement
Figures

Similar articles
-
Regulation of autophagy through SnRK1 and TOR signaling pathways.Plant Signal Behav. 2017 Dec 2;12(12):e1395128. doi: 10.1080/15592324.2017.1395128. Plant Signal Behav. 2017. PMID: 29058995 Free PMC article.
-
Autophagy contributes to positive feedback regulation of SnRK1 signaling in plants.Autophagy. 2023 Dec;19(12):3248-3250. doi: 10.1080/15548627.2023.2247741. Epub 2023 Aug 20. Autophagy. 2023. PMID: 37584544 Free PMC article.
-
Genetic Analyses of the Arabidopsis ATG1 Kinase Complex Reveal Both Kinase-Dependent and Independent Autophagic Routes during Fixed-Carbon Starvation.Plant Cell. 2019 Dec;31(12):2973-2995. doi: 10.1105/tpc.19.00066. Epub 2019 Oct 15. Plant Cell. 2019. PMID: 31615848 Free PMC article.
-
Compartmentalization, a key mechanism controlling the multitasking role of the SnRK1 complex.J Exp Bot. 2022 Nov 15;73(20):7055-7067. doi: 10.1093/jxb/erac315. J Exp Bot. 2022. PMID: 35861169 Free PMC article. Review.
-
SnRK1 and TOR: modulating growth-defense trade-offs in plant stress responses.J Exp Bot. 2019 Apr 15;70(8):2261-2274. doi: 10.1093/jxb/erz066. J Exp Bot. 2019. PMID: 30793201 Review.
Cited by
-
14-3-3 proteins contribute to autophagy by modulating SINAT-mediated degradation of ATG13.Plant Cell. 2022 Nov 29;34(12):4857-4876. doi: 10.1093/plcell/koac273. Plant Cell. 2022. PMID: 36053201 Free PMC article.
-
Signal Integration in Plant Abiotic Stress Responses via Multistep Phosphorelay Signaling.Front Plant Sci. 2021 Feb 17;12:644823. doi: 10.3389/fpls.2021.644823. eCollection 2021. Front Plant Sci. 2021. PMID: 33679861 Free PMC article. Review.
-
Plant Autophagy: An Intricate Process Controlled by Various Signaling Pathways.Front Plant Sci. 2021 Sep 23;12:754982. doi: 10.3389/fpls.2021.754982. eCollection 2021. Front Plant Sci. 2021. PMID: 34630498 Free PMC article. Review.
-
Autophagy in Plants: Both a Puppet and a Puppet Master of Sugars.Front Plant Sci. 2019 Jan 22;10:14. doi: 10.3389/fpls.2019.00014. eCollection 2019. Front Plant Sci. 2019. PMID: 30723485 Free PMC article. Review.
-
Target of rapamycin (TOR) regulates the response to low nitrogen stress via autophagy and hormone pathways in Malus hupehensis.Hortic Res. 2022 Jun 27;9:uhac143. doi: 10.1093/hr/uhac143. eCollection 2022. Hortic Res. 2022. PMID: 36072834 Free PMC article.
References
-
- Yang X, Bassham DC. New Insight into the Mechanism and Function of Autophagy in Plant Cells. Int Rev Cell Mol Biol. 2015;320:1–40. doi: 10.1016/bs.ircmb.2015.07.005 - DOI - PubMed
-
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1–2):169–74. - PubMed
-
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349(2):275–80. - PubMed
-
- Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb Cell. 2016;3(12):588–96. doi: 10.15698/mic2016.12.546 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

