Repliscan: a tool for classifying replication timing regions
- PMID: 28784090
- PMCID: PMC5547489
- DOI: 10.1186/s12859-017-1774-x
Repliscan: a tool for classifying replication timing regions
Abstract
Background: Replication timing experiments that use label incorporation and high throughput sequencing produce peaked data similar to ChIP-Seq experiments. However, the differences in experimental design, coverage density, and possible results make traditional ChIP-Seq analysis methods inappropriate for use with replication timing.
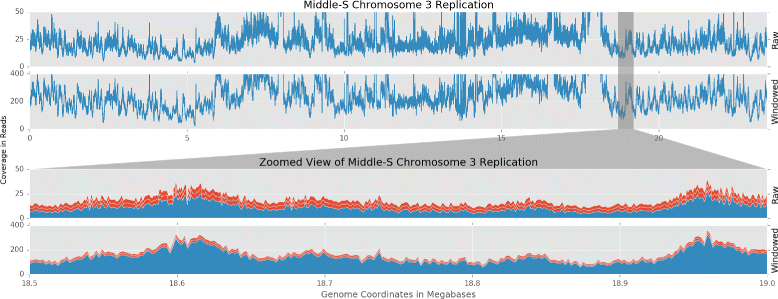
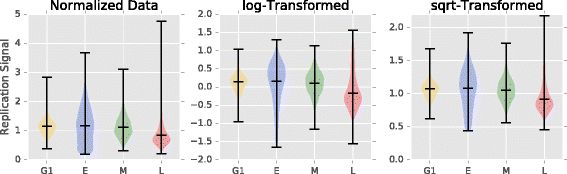
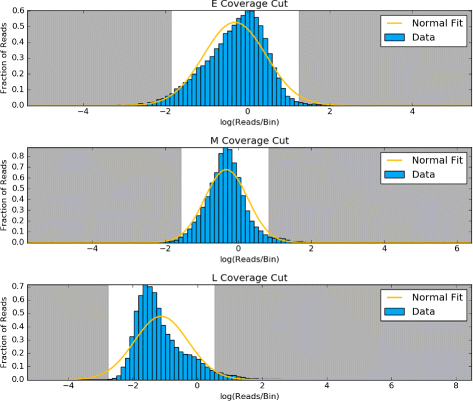
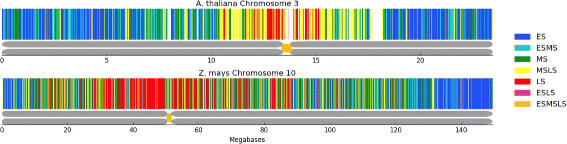
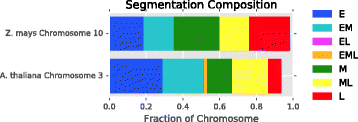
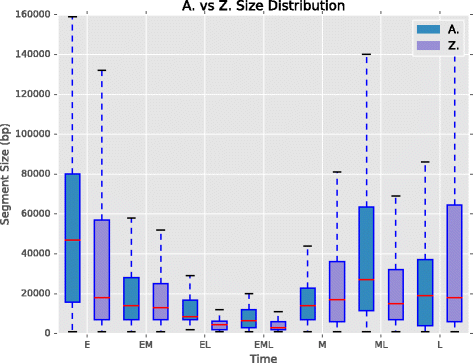
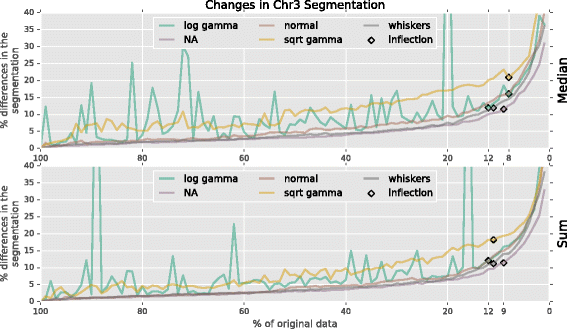
Results: To accurately detect and classify regions of replication across the genome, we present Repliscan. Repliscan robustly normalizes, automatically removes outlying and uninformative data points, and classifies Repli-seq signals into discrete combinations of replication signatures. The quality control steps and self-fitting methods make Repliscan generally applicable and more robust than previous methods that classify regions based on thresholds.
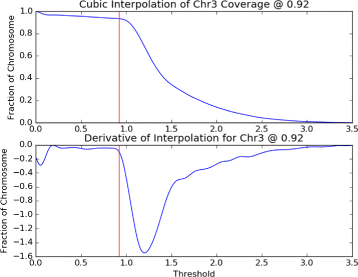
Conclusions: Repliscan is simple and effective to use on organisms with different genome sizes. Even with analysis window sizes as small as 1 kilobase, reliable profiles can be generated with as little as 2.4x coverage.
Keywords: Classification; DNA replication; Repli-seq.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science; 2002.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

