Quantifying enzyme activity in living cells
- PMID: 28784664
- PMCID: PMC5612114
- DOI: 10.1074/jbc.M117.792119
Quantifying enzyme activity in living cells
Abstract
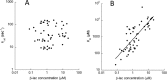
For over a century, enzymatic activity has been studied in vitro, assuming similar activity in the crowded cellular milieu. Here, we determined in real time the catalytic activity of TEM1-β-lactamase inside living cells and compared the values to those obtained in vitro We found the apparent in vivo catalytic efficiency, kcat/Km , to be lower than in vitro, with significant cell-to-cell variability. Surprisingly, the results show that inside the cell the apparent catalytic efficiency decreases, and Km increases with increasing enzyme concentration. To rationalize these findings, we measured enzyme and substrate diffusion rates in the cell and found the latter to be slower than expected. Simulations showed that for attenuated diffusion the substrate flux becomes rate-limiting, explaining why reaction rates in vivo can be independent on enzyme concentrations. The octanol/water partition of the substrate is 4.5, which is in the range of Food and Drug Administration-approved drugs. This suggests substrate-limited reaction rates to be common. These findings indicate that in vitro data cannot be simply extrapolated to the crowded in vivo environment.
Keywords: Michaelis–Menten; beta-lactamase; biophysics; enzyme; enzyme kinetics; in vivo imaging.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures








References
-
- Michaelis L., and Menten M. L. (1913) Die kinetik der invertinwirkung. Biochem. Z. 49, 333–369
-
- Deichmann U., Schuster S., Mazat J. P., and Cornish-Bowden A. (2014) Commemorating the 1913 Michaelis–Menten paper die kinetik der invertinwirkung: three perspectives. FEBS J. 281, 435–463 - PubMed
-
- Fersht A. R. (1999) Structure and Mechanism in Protein Science, W. H. Freeman, New York
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

