Gle1 Regulates RNA Binding of the DEAD-Box Helicase Ded1 in Its Complex Role in Translation Initiation
- PMID: 28784717
- PMCID: PMC5640818
- DOI: 10.1128/MCB.00139-17
Gle1 Regulates RNA Binding of the DEAD-Box Helicase Ded1 in Its Complex Role in Translation Initiation
Abstract
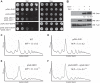
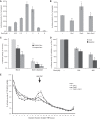
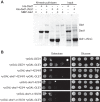
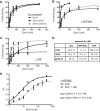
DEAD-box proteins (DBPs) are required in gene expression to facilitate changes to ribonucleoprotein complexes, but the cellular mechanisms and regulation of DBPs are not fully defined. Gle1 is a multifunctional regulator of DBPs with roles in mRNA export and translation. In translation, Gle1 modulates Ded1, a DBP required for initiation. However, DED1 overexpression causes defects, suggesting that Ded1 can promote or repress translation in different contexts. Here we show that GLE1 expression suppresses the repressive effects of DED1 in vivo and Gle1 counteracts Ded1 in translation assays in vitro Furthermore, both Ded1 and Gle1 affect the assembly of preinitiation complexes. Through mutation analysis and binding assays, we show that Gle1 inhibits Ded1 by reducing its affinity for RNA. Our results are consistent with a model wherein active Ded1 promotes translation but inactive or excess Ded1 leads to translation repression. Gle1 can inhibit either role of Ded1, positioning it as a gatekeeper to optimize Ded1 activity to the appropriate level for translation. This study suggests a paradigm for finely controlling the activity of DEAD-box proteins to optimize their function in RNA-based processes. It also positions the versatile regulator Gle1 as a potential node for the coordination of different steps of gene expression.
Keywords: DEAD box; RNA; helicase; nuclear export; translation; yeast.
Copyright © 2017 American Society for Microbiology.
Figures

References
-
- Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, Milde T, Bourdeaut F, Ryzhova M, Sturm D, Pfaff E, Stark S, Hutter S, Seker-Cin H, Johann P, Bender S, Schmidt C, Rausch T, Shih D, Reimand J, Sieber L, Wittmann A, Linke L, Witt H, Weber UD, Zapatka M, Konig R, Beroukhim R, Bergthold G, van Sluis P, Volckmann R, Koster J, Versteeg R, Schmidt S, Wolf S, Lawerenz C, Bartholomae CC, von Kalle C, Unterberg A, Herold-Mende C, Hofer S, Kulozik AE, von Deimling A, Scheurlen W, Felsberg J, Reifenberger G, Hasselblatt M, Crawford JR, Grant GA, Jabado N, Perry A, Cowdrey C, Croul S, Zadeh G, Korbel JO, Doz F, Delattre O, Bader GD, McCabe MG, Collins VP, Kieran MW, Cho YJ, Pomeroy SL, Witt O, Brors B, Taylor MD, Schüller U, Korshunov A, Eils R, Wechsler-Reya RJ, Lichter P, Pfister SM, ICGC PedBrain Tumor Project . 2014. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 25:393–405. doi:10.1016/j.ccr.2014.02.004. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
