Molecularly targeted drug combinations demonstrate selective effectiveness for myeloid- and lymphoid-derived hematologic malignancies
- PMID: 28784769
- PMCID: PMC5594650
- DOI: 10.1073/pnas.1703094114
Molecularly targeted drug combinations demonstrate selective effectiveness for myeloid- and lymphoid-derived hematologic malignancies
Abstract
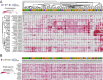
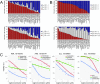
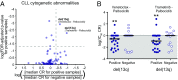
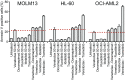
Translating the genetic and epigenetic heterogeneity underlying human cancers into therapeutic strategies is an ongoing challenge. Large-scale sequencing efforts have uncovered a spectrum of mutations in many hematologic malignancies, including acute myeloid leukemia (AML), suggesting that combinations of agents will be required to treat these diseases effectively. Combinatorial approaches will also be critical for combating the emergence of genetically heterogeneous subclones, rescue signals in the microenvironment, and tumor-intrinsic feedback pathways that all contribute to disease relapse. To identify novel and effective drug combinations, we performed ex vivo sensitivity profiling of 122 primary patient samples from a variety of hematologic malignancies against a panel of 48 drug combinations. The combinations were designed as drug pairs that target nonoverlapping biological pathways and comprise drugs from different classes, preferably with Food and Drug Administration approval. A combination ratio (CR) was derived for each drug pair, and CRs were evaluated with respect to diagnostic categories as well as against genetic, cytogenetic, and cellular phenotypes of specimens from the two largest disease categories: AML and chronic lymphocytic leukemia (CLL). Nearly all tested combinations involving a BCL2 inhibitor showed additional benefit in patients with myeloid malignancies, whereas select combinations involving PI3K, CSF1R, or bromodomain inhibitors showed preferential benefit in lymphoid malignancies. Expanded analyses of patients with AML and CLL revealed specific patterns of ex vivo drug combination efficacy that were associated with select genetic, cytogenetic, and phenotypic disease subsets, warranting further evaluation. These findings highlight the heuristic value of an integrated functional genomic approach to the identification of novel treatment strategies for hematologic malignancies.
Keywords: drug combinations; ex vivo assay; hematologic malignancies; targeted therapies.
Conflict of interest statement
Conflict of interest statement: D.A.P. serves on the advisory boards for Pharmacyclics and Gilead. J.W.T. receives research support from Agios, Array Biopharma, Aptose, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen R&D, Seattle Genetics, Syros, and Takeda and is a consultant for Leap Oncology. B.J.D. serves on the advisory boards for Gilead and Roche TCRC. B.J.D. is principal investigator or coinvestigator on Novartis and BMS clinical trials. His institution, Oregon Health & Science University, has contracts with these companies to pay for patient costs, nurse and data manager salaries, and institutional overhead. He does not derive salary, nor does his laboratory receive funds from these contracts. M.W.D. serves on the advisory boards and/or as a consultant for Novartis, Incyte, and BMS and receives research funding from BMS and Gilead. The authors certify that all compounds and combinations tested in this study were chosen without input from any of our industry partners.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

