Opsin-Mediated Inhibition of Bacterioruberin Synthesis in Halophilic Archaea
- PMID: 28784816
- PMCID: PMC5626960
- DOI: 10.1128/JB.00303-17
Opsin-Mediated Inhibition of Bacterioruberin Synthesis in Halophilic Archaea
Abstract
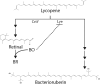
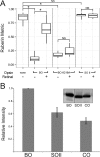
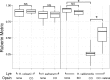
Halophilic archaea often inhabit environments with limited oxygen, and many produce ion-pumping rhodopsin complexes that allow them to maintain electrochemical gradients when aerobic respiration is inhibited. Rhodopsins require a protein, an opsin, and an organic cofactor, retinal. We previously demonstrated that in Halobacterium salinarum, bacterioopsin (BO), when not bound by retinal, inhibits the production of bacterioruberin, a biochemical pathway that shares intermediates with retinal biosynthesis. In this work, we used heterologous expression in a related halophilic archaeon, Haloferax volcanii, to demonstrate that BO is sufficient to inhibit bacterioruberin synthesis catalyzed by the H. salinarum lycopene elongase (Lye) enzyme. This inhibition was observed both in liquid culture and in a novel colorimetric assay to quantify bacterioruberin abundance based on the colony color. Addition of retinal to convert BO to the bacteriorhodopsin complex resulted in a partial rescue of bacterioruberin production. To explore if this regulatory mechanism occurs in other organisms, we expressed a Lye homolog and an opsin from Haloarcula vallismortis in H. volcaniiH. vallismortis cruxopsin-3 expression inhibited bacterioruberin synthesis catalyzed by H. vallismortis Lye but had no effect when bacterioruberin synthesis was catalyzed by H. salinarum or H. volcanii Lye. Conversely, H. salinarum BO did not inhibit H. vallismortis Lye activity. Together, our data suggest that opsin-mediated inhibition of Lye is potentially widespread and represents an elegant regulatory mechanism that allows organisms to efficiently utilize ion-pumping rhodopsins obtained through lateral gene transfer.IMPORTANCE Many enzymes are complexes of proteins and nonprotein organic molecules called cofactors. To ensure efficient formation of functional complexes, organisms must regulate the production of proteins and cofactors. To study this regulation, we used bacteriorhodopsin from the archaeon Halobacterium salinarum Bacteriorhodopsin consists of the bacterioopsin protein and a retinal cofactor. In this article, we further characterize a novel regulatory mechanism in which bacterioopsin promotes retinal production by inhibiting a reaction that consumes lycopene, a retinal precursor. By expressing H. salinarum genes in a different organism, Haloferax volcanii, we demonstrated that bacterioopsin alone is sufficient for this inhibition. We also found that an opsin from Haloarcula vallismortis has inhibitory activity, suggesting that this regulatory mechanism might be found in other organisms.
Keywords: C50 carotenoid; UbiA prenyltransferase; carotenoid biosynthesis; cofactor biosynthesis; membrane protein biogenesis; microbial rhodopsin; proteorhodopsin.
Copyright © 2017 American Society for Microbiology.
Figures



Similar articles
-
Species Widely Distributed in Halophilic Archaea Exhibit Opsin-Mediated Inhibition of Bacterioruberin Biosynthesis.J Bacteriol. 2018 Dec 20;201(2):e00576-18. doi: 10.1128/JB.00576-18. Print 2019 Jan 15. J Bacteriol. 2018. PMID: 30373756 Free PMC article.
-
Bacterioopsin-mediated regulation of bacterioruberin biosynthesis in Halobacterium salinarum.J Bacteriol. 2011 Oct;193(20):5658-67. doi: 10.1128/JB.05376-11. Epub 2011 Aug 12. J Bacteriol. 2011. PMID: 21840984 Free PMC article.
-
Complete biosynthetic pathway of the C50 carotenoid bacterioruberin from lycopene in the extremely halophilic archaeon Haloarcula japonica.J Bacteriol. 2015 May;197(9):1614-23. doi: 10.1128/JB.02523-14. Epub 2015 Feb 23. J Bacteriol. 2015. PMID: 25712483 Free PMC article.
-
Variations on a molecular switch: transport and sensory signalling by archaeal rhodopsins.Mol Microbiol. 1998 Jun;28(6):1051-8. doi: 10.1046/j.1365-2958.1998.00859.x. Mol Microbiol. 1998. PMID: 9680197 Review.
-
The structure and mechanism of the family of retinal proteins from halophilic archaea.Curr Opin Struct Biol. 1998 Aug;8(4):489-500. doi: 10.1016/s0959-440x(98)80128-0. Curr Opin Struct Biol. 1998. PMID: 9729742 Review.
Cited by
-
Proteolysis at the Archaeal Membrane: Advances on the Biological Function and Natural Targets of Membrane-Localized Proteases in Haloferax volcanii.Front Microbiol. 2022 Jun 24;13:940865. doi: 10.3389/fmicb.2022.940865. eCollection 2022. Front Microbiol. 2022. PMID: 35814708 Free PMC article. Review.
-
Species Widely Distributed in Halophilic Archaea Exhibit Opsin-Mediated Inhibition of Bacterioruberin Biosynthesis.J Bacteriol. 2018 Dec 20;201(2):e00576-18. doi: 10.1128/JB.00576-18. Print 2019 Jan 15. J Bacteriol. 2018. PMID: 30373756 Free PMC article.
-
Carotenoids as a Protection Mechanism against Oxidative Stress in Haloferax mediterranei.Antioxidants (Basel). 2020 Oct 29;9(11):1060. doi: 10.3390/antiox9111060. Antioxidants (Basel). 2020. PMID: 33137984 Free PMC article.
-
Archaea Carotenoids: Natural Pigments with Unexplored Innovative Potential.Mar Drugs. 2022 Aug 17;20(8):524. doi: 10.3390/md20080524. Mar Drugs. 2022. PMID: 36005527 Free PMC article. Review.
-
Cellular differentiation into hyphae and spores in halophilic archaea.Nat Commun. 2023 Apr 1;14(1):1827. doi: 10.1038/s41467-023-37389-w. Nat Commun. 2023. PMID: 37005419 Free PMC article.
References
-
- Oesterhelt D, Stoeckenius W. 1971. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol 233:149–152. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

