α3-Deletion Isoform of HLA-A11 Modulates Cytotoxicity of NK Cells: Correlations with HIV-1 Infection of Cells
- PMID: 28784847
- PMCID: PMC5583747
- DOI: 10.4049/jimmunol.1602183
α3-Deletion Isoform of HLA-A11 Modulates Cytotoxicity of NK Cells: Correlations with HIV-1 Infection of Cells
Abstract
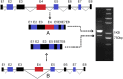
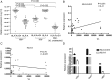
Alternative splicing occurs frequently in many genes, especially those involved in immunity. Unfortunately, the functions of many alternatively spliced molecules from immunologically relevant genes remain unknown. Classical HLA-I molecules are expressed on almost all nucleated cells and play a pivotal role in both innate and adaptive immunity. Although splice variants of HLA-I genes have been reported, the details of their functions have not been reported. In the current study, we determined the characteristics, expression, and function of a novel splice variant of HLA-A11 named HLA-A11svE4 HLA-A11svE4 is located on the cell surface without β2-microglobulin (β2m). Additionally, HLA-A11svE4 forms homodimers as well as heterodimers with HLA-A open conformers, instead of combining with β2m. Moreover, HLA-A11svE4 inhibits the activation of NK cells to protect target cells. Compared with β2m and HLA-A11, the heterodimer of HLA-A11svE4 and HLA-A11 protected target cells from lysis by NK cells more effectively. Furthermore, HLA-AsvE4 expression was upregulated by HIV-1 in vivo and by HSV, CMV, and hepatitis B virus in vitro. In addition, our findings indicated that HLA-A11svE4 molecules were functional in activating CD8+ T cells through Ag presentation. Taken together, these results suggested that HLA-A11svE4 can homodimerize and form a novel heterodimeric complex with HLA-A11 open conformers. Furthermore, the data are consistent with HLA-A11svE4 playing a role in the immune escape of HIV-1.
Copyright © 2017 by The American Association of Immunologists, Inc.
Figures







References
-
- Raghavan S., Selvaraj P., Swaminathan S., Narendran G. 2009. Short communication: association of HLA-A*1101 with resistance and B*4006 with susceptibility to HIV and HIV-TB: an in silico analysis of promiscuous T cell epitopes. AIDS Res. Hum. Retroviruses 25: 1023–1028. - PubMed
-
- Li L., Bouvier M. 2004. Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue. J. Immunol. 172: 6175–6184. - PubMed
-
- Harrer T., Harrer E., Barbosa P., Kaufmann F., Wagner R., Brüggemann S., Kalden J. R., Feinberg M., Johnson R. P., Buchbinder S., Walker B. D. 1998. Recognition of two overlapping CTL epitopes in HIV-1 p17 by CTL from a long-term nonprogressing HIV-1-infected individual. J. Immunol. 161: 4875–4881. - PubMed
-
- Beyrer C., Artenstein A. W., Rugpao S., Stephens H., VanCott T. C., Robb M. L., Rinkaew M., Birx D. L., Khamboonruang C., Zimmerman P. A., et al. Chiang Mai HEPS Working Group 1999. Epidemiologic and biologic characterization of a cohort of human immunodeficiency virus type 1 highly exposed, persistently seronegative female sex workers in northern Thailand. J. Infect. Dis. 179: 59–67. - PubMed
-
- Sriwanthana B., Hodge T., Mastro T. D., Dezzutti C. S., Bond K., Stephens H. A., Kostrikis L. G., Limpakarnjanarat K., Young N. L., Qari S. H., et al. 2001. HIV-specific cytotoxic T lymphocytes, HLA-A11, and chemokine-related factors may act synergistically to determine HIV resistance in CCR5 delta32-negative female sex workers in Chiang Rai, northern Thailand. AIDS Res. Hum. Retroviruses 17: 719–734. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

