Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment
- PMID: 28784993
- PMCID: PMC5547136
- DOI: 10.1038/s41598-017-07713-8
Carriage frequency, phenotypic, and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolated from dental health-care personnel, patients, and environment
Abstract
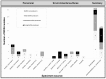
There is limited data on methicillin-resistant Staphylococcus aureus (MRSA) carriage in dental clinics. 1300 specimens from patients, health personnel, and environmental surfaces of a dental clinic in Egypt were tested for MRSA. Antibiotic susceptibility, biofilm formation, Staphylococcal protein A (spa) typing, SCCmec typing, and PCR-based assays were used to detect mecA, mecC, vanA, Panton-Valentine Leukocidin toxin (PVL), and toxic shock syndrome toxin-1 (tst) genes. Among 34 mecA-positive MRSA isolates, five (14.7%) were PVL-positive, seventeen (50%) were tst-positive, ten (29.4%) were vanA-positive, while none harboured mecC. MRSA hand carriage rates in patients, nurses, and dentists were 9.8%, 6.6%, and 5%. The respective nasal colonization rates were 11.1%, 6.7%, and 9.7%. 1.3% of the environmental isolates were MRSA-positive. Strong and moderate biofilm-forming isolates represented 23.5% and 29.4% of MRSA isolates. 24 MRSA isolates (70.6%) were multi-resistant and 18 (52.9%) harboured SCCmec IV. Among eight spa types, t223 (26.5%), t267 (23.5%), and t14339 (23.5%) were predominant. We noted an alarming genetic relatedness between 7 (20.6%) MRSA isolates and the epidemic EMRSA-15 clone, as well as a combined occurrence of tst and PVL in 3 (8.8%) isolates. Results suggest high MRSA pathogenicity in dental wards highlighting the need for more efficient surveillance/infection control strategies.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Prevalence of oxacillin-susceptible methicillin-resistant Staphylococcus aureus nasal carriage and their clonal diversity among patients attending public health-care facilities.Indian J Med Microbiol. 2020 Apr-Jun;38(2):192-199. doi: 10.4103/ijmm.IJMM_20_157. Indian J Med Microbiol. 2020. PMID: 32883933
-
Detection of Panton-Valentine Leukocidin-Positive Methicillin-Resistant Staphylococcus aureus Nasal Carriage among Egyptian Health Care Workers.Surg Infect (Larchmt). 2016 Jun;17(3):369-75. doi: 10.1089/sur.2015.192. Epub 2016 Mar 16. Surg Infect (Larchmt). 2016. PMID: 26983032
-
Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt.Braz J Infect Dis. 2015 Jan-Feb;19(1):68-76. doi: 10.1016/j.bjid.2014.09.005. Epub 2014 Dec 15. Braz J Infect Dis. 2015. PMID: 25523075 Free PMC article.
-
Molecular characterization and genotyping of methicillin-resistant Staphylococcus aureus in nasal carriage of healthy Iranian children.J Med Microbiol. 2019 Mar;68(3):374-378. doi: 10.1099/jmm.0.000924. Epub 2019 Jan 30. J Med Microbiol. 2019. PMID: 30698518
-
Global prevalence and dynamics of mecA and mecC genes in MRSA: Meta-meta-analysis, meta-regression, and temporal investigation.J Infect Public Health. 2025 Jul;18(7):102802. doi: 10.1016/j.jiph.2025.102802. Epub 2025 Apr 30. J Infect Public Health. 2025. PMID: 40319833 Review.
Cited by
-
Staphylococcus aureus Isolated from the Oral Cavity: Phage Susceptibility in Relation to Antibiotic Resistance.Antibiotics (Basel). 2021 Oct 31;10(11):1329. doi: 10.3390/antibiotics10111329. Antibiotics (Basel). 2021. PMID: 34827268 Free PMC article.
-
Isolation and Molecular Identification of Virulence, Antimicrobial and Heavy Metal Resistance Genes in Livestock-Associated Methicillin-Resistant Staphylococcus aureus.Pathogens. 2019 Jun 14;8(2):79. doi: 10.3390/pathogens8020079. Pathogens. 2019. PMID: 31207959 Free PMC article.
-
Nosocomial Infections: Do Not Forget the Parasites!Pathogens. 2021 Feb 19;10(2):238. doi: 10.3390/pathogens10020238. Pathogens. 2021. PMID: 33669761 Free PMC article. Review.
-
Phenotype-Genotype Characterization and Antibiotic-Resistance Correlations Among Colonizing and Infectious Methicillin-Resistant Staphylococcus aureus Recovered from Intensive Care Units.Infect Drug Resist. 2021 Apr 21;14:1557-1571. doi: 10.2147/IDR.S296000. eCollection 2021. Infect Drug Resist. 2021. PMID: 33907431 Free PMC article.
-
Methicillin-Resistant Staphylococcus aureus Harboring mecC Still Eludes Us in East London, United Kingdom.J Clin Microbiol. 2019 May 24;57(6):e00020-19. doi: 10.1128/JCM.00020-19. Print 2019 Jun. J Clin Microbiol. 2019. PMID: 30971461 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

