Isolation and Characterization of phiLLS, a Novel Phage with Potential Biocontrol Agent against Multidrug-Resistant Escherichia coli
- PMID: 28785246
- PMCID: PMC5519627
- DOI: 10.3389/fmicb.2017.01355
Isolation and Characterization of phiLLS, a Novel Phage with Potential Biocontrol Agent against Multidrug-Resistant Escherichia coli
Abstract
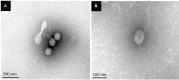
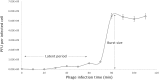
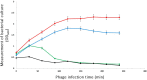
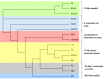
Foodborne diseases are a serious and growing problem, and the incidence and prevalence of antimicrobial resistance among foodborne pathogens is reported to have increased. The emergence of antibiotic-resistant bacterial strains demands novel strategies to counteract this epidemic. In this regard, lytic bacteriophages have reemerged as an alternative for the control of pathogenic bacteria. However, the effective use of phages relies on appropriate biological and genomic characterization. In this study, we present the isolation and characterization of a novel bacteriophage named phiLLS, which has shown strong lytic activity against generic and multidrug-resistant Escherichia coli strains. Transmission electron microscopy of phiLLS morphology revealed that it belongs to the Siphoviridae family. Furthermore, this phage exhibited a relatively large burst size of 176 plaque-forming units per infected cell. Phage phiLLS significantly reduced the growth of E. coli under laboratory conditions. Analyses of restriction profiles showed the presence of submolar fragments, confirming that phiLLS is a pac-type phage. Phylogenetic analysis based on the amino acid sequence of large terminase subunits confirmed that this phage uses a headful packaging strategy to package their genome. Genomic sequencing and bioinformatic analysis showed that phiLLS is a novel bacteriophage that is most closely related to T5-like phages. In silico analysis indicated that the phiLLS genome consists of 107,263 bp (39.0 % GC content) encoding 160 putative ORFs, 16 tRNAs, several potential promoters and transcriptional terminators. Genome analysis suggests that the phage phiLLS is strictly lytic without carrying genes associated with virulence factors and/or potential immunoreactive allergen proteins. The bacteriophage isolated in this study has shown promising results in the biocontrol of bacterial growth under in vitro conditions, suggesting that it may prove useful as an alternative agent for the control of foodborne pathogens. However, further oral toxicity testing is needed to ensure the safety of phage use.
Keywords: bacteriophage phiLLS; biotechnological applications; genome sequence; in silico.
Figures

Similar articles
-
Characterization and complete genome sequence analysis of a newly isolatedphage against Vibrio parahaemolyticus from sick shrimp in Qingdao, China.PLoS One. 2022 May 4;17(5):e0266683. doi: 10.1371/journal.pone.0266683. eCollection 2022. PLoS One. 2022. PMID: 35507581 Free PMC article.
-
Genomic and Proteomic Characterizations of Sfin-1, a Novel Lytic Phage Infecting Multidrug-Resistant Shigella spp. and Escherichia coli C.Front Microbiol. 2019 Aug 22;10:1876. doi: 10.3389/fmicb.2019.01876. eCollection 2019. Front Microbiol. 2019. PMID: 31507544 Free PMC article.
-
Isolation, characterization and genomic analysis of a novel phage IME178 with lytic activity against Escherichia coli.Microb Pathog. 2023 Jun;179:106099. doi: 10.1016/j.micpath.2023.106099. Epub 2023 Apr 14. Microb Pathog. 2023. PMID: 37060965
-
A lytic phage to control multidrug-resistant avian pathogenic Escherichia coli (APEC) infection.Front Cell Infect Microbiol. 2023 Sep 7;13:1253815. doi: 10.3389/fcimb.2023.1253815. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37743864 Free PMC article.
-
Characterization of novel bacteriophage phiC119 capable of lysing multidrug-resistant Shiga toxin-producing Escherichia coli O157:H7.PeerJ. 2016 Sep 13;4:e2423. doi: 10.7717/peerj.2423. eCollection 2016. PeerJ. 2016. PMID: 27672499 Free PMC article.
Cited by
-
Characterization of vB_VpaP_MGD2, a newly isolated bacteriophage with biocontrol potential against multidrug-resistant Vibrio parahaemolyticus.Arch Virol. 2021 Feb;166(2):413-426. doi: 10.1007/s00705-020-04887-x. Epub 2021 Jan 3. Arch Virol. 2021. PMID: 33389104
-
A New Casjensviridae Bacteriophage Isolated from Hospital Sewage for Inactivation of Biofilms of Carbapenem Resistant Klebsiella pneumoniae Clinical Isolates.Pharmaceutics. 2024 Jul 5;16(7):904. doi: 10.3390/pharmaceutics16070904. Pharmaceutics. 2024. PMID: 39065601 Free PMC article.
-
Characterization and complete genome sequence analysis of a newly isolatedphage against Vibrio parahaemolyticus from sick shrimp in Qingdao, China.PLoS One. 2022 May 4;17(5):e0266683. doi: 10.1371/journal.pone.0266683. eCollection 2022. PLoS One. 2022. PMID: 35507581 Free PMC article.
-
Characterization and Genome Analysis of Vibrio campbellii Lytic Bacteriophage OPA17.Microbiol Spectr. 2023 Jan 31;11(2):e0162322. doi: 10.1128/spectrum.01623-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36719217 Free PMC article.
-
Lytic KFS-SE2 phage as a novel bio-receptor for Salmonella Enteritidis detection.J Microbiol. 2019 Feb;57(2):170-179. doi: 10.1007/s12275-019-8610-0. Epub 2019 Jan 31. J Microbiol. 2019. PMID: 30706346
References
-
- Amézquita-López B. A., Quiñones B., Soto-Beltrán M., Lee B. G., Yambao J. C., Lugo-Melchor O. Y., et al. . (2016). Antimicrobial resistance profiles of Shiga toxin-producing Escherichia coli O157 and Non-O157 recovered from domestic farm animals in rural communities in Northwestern Mexico. Antimicrobial. Resist. Infect. Control 5:1. 10.1186/s13756-015-0100-5 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

