Primary Cilia Signaling Shapes the Development of Interneuronal Connectivity
- PMID: 28787594
- PMCID: PMC5571900
- DOI: 10.1016/j.devcel.2017.07.010
Primary Cilia Signaling Shapes the Development of Interneuronal Connectivity
Abstract
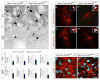
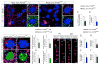
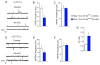
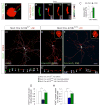
Appropriate growth and synaptic integration of GABAergic inhibitory interneurons are essential for functional neural circuits in the brain. Here, we demonstrate that disruption of primary cilia function following the selective loss of ciliary GTPase Arl13b in interneurons impairs interneuronal morphology and synaptic connectivity, leading to altered excitatory/inhibitory activity balance. The altered morphology and connectivity of cilia mutant interneurons and the functional deficits are rescued by either chemogenetic activation of ciliary G-protein-coupled receptor (GPCR) signaling or the selective induction of Sstr3, a ciliary GPCR, in Arl13b-deficient cilia. Our results thus define a specific requirement for primary cilia-mediated GPCR signaling in interneuronal connectivity and inhibitory circuit formation.
Keywords: Arl13b; GPCR signaling; Joubert syndrome related disorders; autism spectrum disorders; ciliopathies; circuitry; interneurons; primary cilia.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


References
-
- Adamantidis A, Thomas E, Foidart A, Tyhon A, Coumans B, Minet A, Tirelli E, Seutin V, Grisar T, Lakaye B. Disrupting the melanin-concentrating hormone receptor 1 in mice leads to cognitive deficits and alterations of NMDA receptor function. The European journal of neuroscience. 2005;21:2837–2844. - PubMed
-
- Amann-Zalcenstein D, Avidan N, Kanyas K, Ebstein RP, Kohn Y, Hamdan A, Ben-Asher E, Karni O, Mujaheed M, Segman RH, et al. AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia. European journal of human genetics: EJHG. 2006;14:1111–1119. - PubMed
-
- Baraban SC, Southwell DG, Estrada RC, Jones DL, Sebe JY, Alfaro-Cervello C, García-Verdugo JM, Rubenstein JLR, Alvarez-Buylla A. Reduction of seizures by transplantation of cortical GABAergic interneuron precursors into Kv1.1 mutant mice. Proc Natl Acad Sci U S A. 2009;106:15472–15477. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

