Transplanted human fecal microbiota enhanced Guillain Barré syndrome autoantibody responses after Campylobacter jejuni infection in C57BL/6 mice
- PMID: 28789710
- PMCID: PMC5547673
- DOI: 10.1186/s40168-017-0284-4
Transplanted human fecal microbiota enhanced Guillain Barré syndrome autoantibody responses after Campylobacter jejuni infection in C57BL/6 mice
Abstract
Background: Campylobacter jejuni is the leading antecedent infection to the autoimmune neuropathy Guillain-Barré syndrome (GBS), which is accompanied by an autoimmune anti-ganglioside antibody attack on peripheral nerves. Previously, we showed that contrasting immune responses mediate C. jejuni induced colitis and autoimmunity in interleukin-10 (IL-10)-deficient mice, dependent upon the infecting strain. Strains from colitis patients elicited T helper 1 (TH1)-dependent inflammatory responses while strains from GBS patients elicited TH2-dependent autoantibody production. Both syndromes were exacerbated by antibiotic depletion of the microbiota, but other factors controlling susceptibility to GBS are unknown.
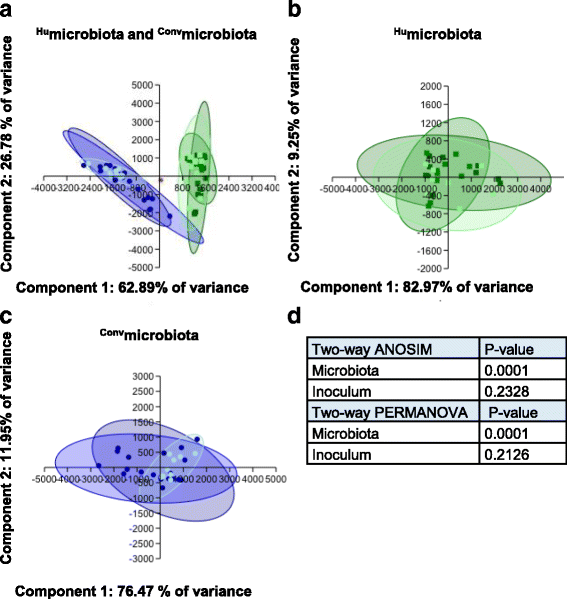
Methods: Using 16S rRNA gene high-throughput sequencing, we examined whether structure of the gut microbial community alters host (1) gastrointestinal inflammation or (2) anti-ganglioside antibody responses after infection with C. jejuni strains from colitis or GBS patients. We compared these responses in C57BL/6 mice with either (1) stable human gut microbiota (Humicrobiota) transplants or (2) conventional mouse microbiota (Convmicrobiota).
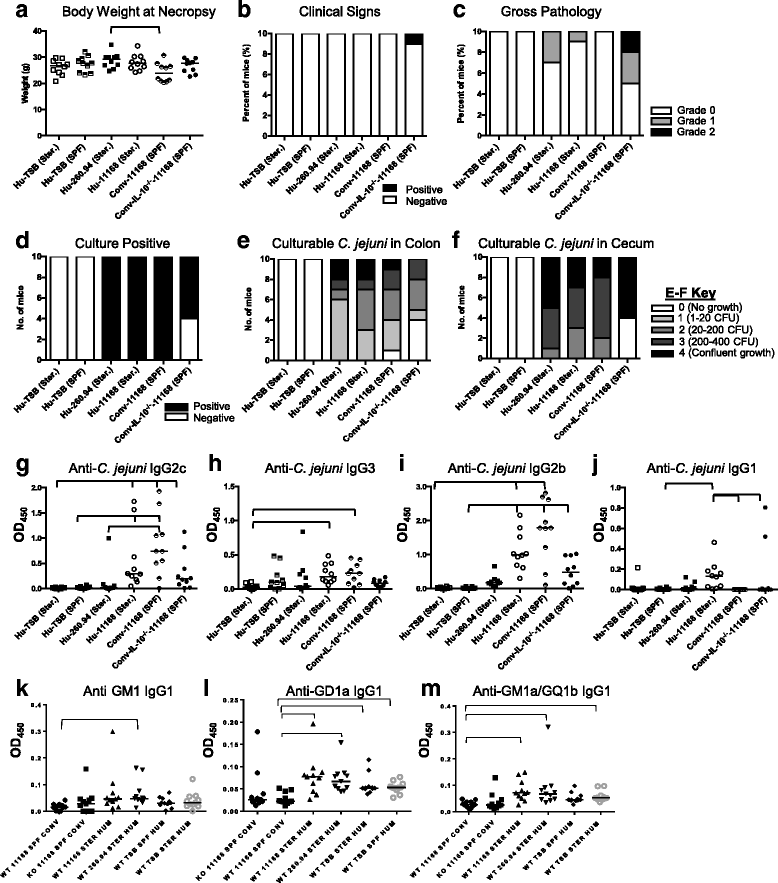
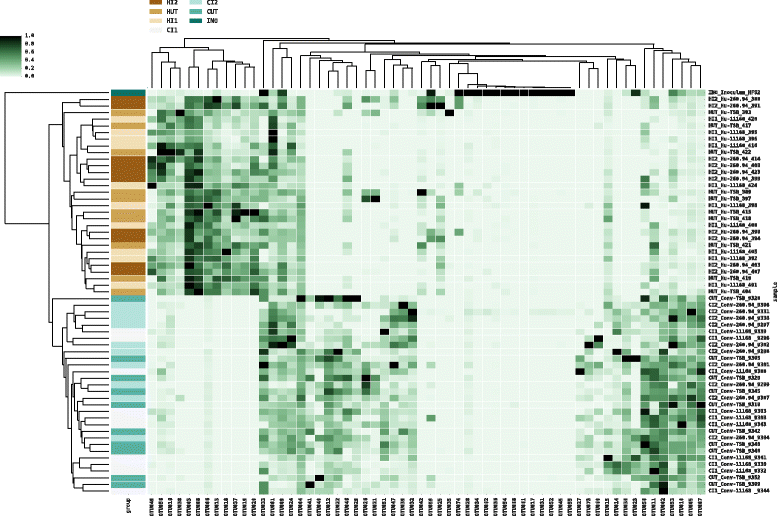
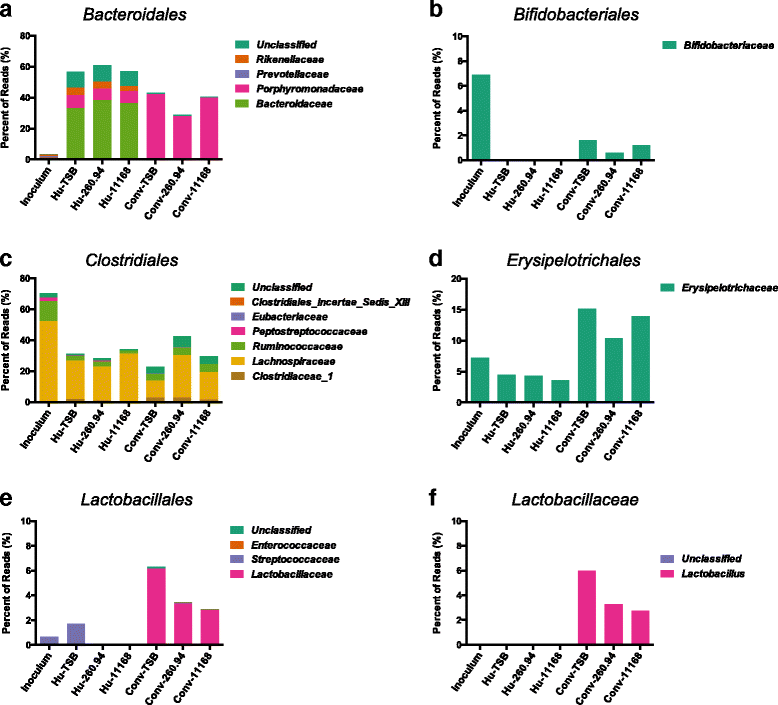
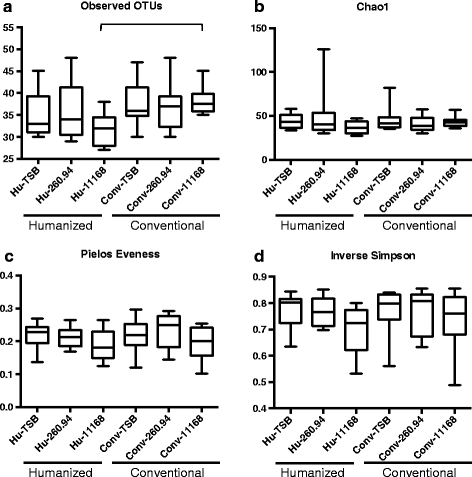
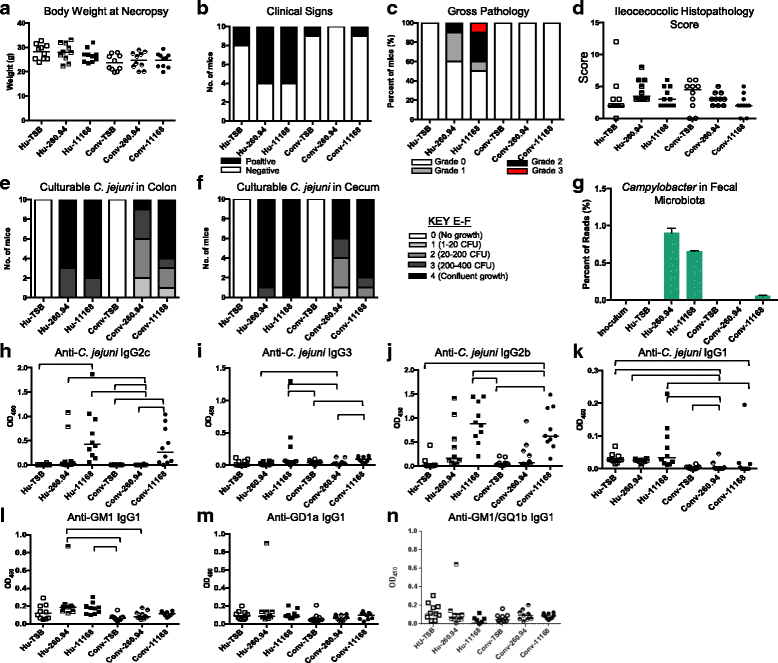
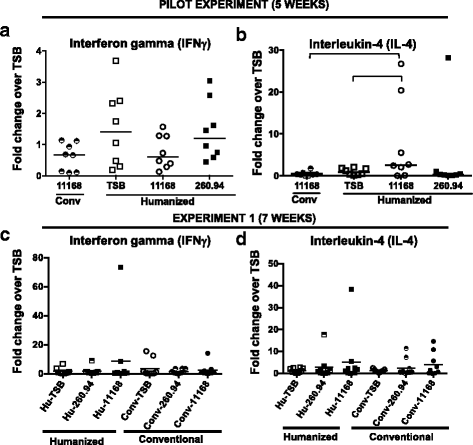
Results: Inoculating germ-free C57BL/6 wild-type (WT) mice with a mixed human fecal slurry provided a murine model that stably passed its microbiota over >20 generations. Mice were housed in specific pathogen-free (SPF) facilities, while extra precautions of having caretakers wear sterile garb along with limited access ensured that no mouse pathogens were acquired. Humicrobiota conferred many changes upon the WT model in contrast to previous results, which showed only colonization with no disease after C. jejuni challenge. When compared to Convmicrobiota mice for susceptibility to C. jejuni enteric or GBS patient strains, infected Humicrobiota mice had (1) 10-100 fold increases in C. jejuni colonization of both strains, (2) pathologic change in draining lymph nodes but only mild changes in colon or cecal lamina propria, (3) significantly lower Th1/Th17-dependent anti-C. jejuni responses, (4) significantly higher IL-4 responses at 5 but not 7 weeks post infection (PI), (5) significantly higher Th2-dependent anti-C. jejuni responses, and (6) significantly elevated anti-ganglioside autoantibodies after C. jejuni infection. These responses in Humicrobiota mice were correlated with a dominant Bacteroidetes and Firmicutes microbiota.
Conclusions: These data demonstrate that Humicrobiota altered host-pathogen interactions in infected mice, increasing colonization and Th-2 and autoimmune responses in a C. jejuni strain-dependent manner. Thus, microbiota composition is another factor controlling susceptibility to GBS.
Keywords: Autoimmunity; Broad-spectrum antibiotics; Campylobacter jejuni; Commensal microbiota; Gastrointestinal inflammation; Guillain-Barré syndrome; Mouse models.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

