Pathogenicity Locus, Core Genome, and Accessory Gene Contributions to Clostridium difficile Virulence
- PMID: 28790208
- PMCID: PMC5550754
- DOI: 10.1128/mBio.00885-17
Pathogenicity Locus, Core Genome, and Accessory Gene Contributions to Clostridium difficile Virulence
Abstract
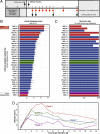
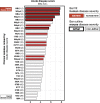
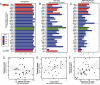
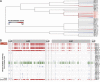
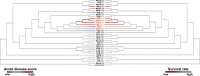
Clostridium difficile is a spore-forming anaerobic bacterium that causes colitis in patients with disrupted colonic microbiota. While some individuals are asymptomatic C. difficile carriers, symptomatic disease ranges from mild diarrhea to potentially lethal toxic megacolon. The wide disease spectrum has been attributed to the infected host's age, underlying diseases, immune status, and microbiome composition. However, strain-specific differences in C. difficile virulence have also been implicated in determining colitis severity. Because patients infected with C. difficile are unique in terms of medical history, microbiome composition, and immune competence, determining the relative contribution of C. difficile virulence to disease severity has been challenging, and conclusions regarding the virulence of specific strains have been inconsistent. To address this, we used a mouse model to test 33 clinical C. difficile strains isolated from patients with disease severities ranging from asymptomatic carriage to severe colitis, and we determined their relative in vivo virulence in genetically identical, antibiotic-pretreated mice. We found that murine infections with C. difficile clade 2 strains (including multilocus sequence type 1/ribotype 027) were associated with higher lethality and that C. difficile strains associated with greater human disease severity caused more severe disease in mice. While toxin production was not strongly correlated with in vivo colonic pathology, the ability of C. difficile strains to grow in the presence of secondary bile acids was associated with greater disease severity. Whole-genome sequencing and identification of core and accessory genes identified a subset of accessory genes that distinguish high-virulence from lower-virulence C. difficile strains.IMPORTANCEClostridium difficile is an important cause of hospital-associated intestinal infections, and recent years have seen an increase in the number and severity of cases in the United States. A patient's antibiotic history, immune status, and medical comorbidities determine, in part, the severity of C. difficile infection. The relative virulence of different clinical C. difficile strains, although postulated to determine disease severity in patients, has been more difficult to consistently associate with mild versus severe colitis. We tested 33 distinct clinical C. difficile isolates for their ability to cause disease in genetically identical mice and found that C. difficile strains belonging to clade 2 were associated with higher mortality. Differences in survival were not attributed to differences in toxin production but likely resulted from the distinct gene content in the various clinical isolates.
Keywords: Clostridium difficile; accessory genome; bile salt; genome analysis; pathogenicity locus; toxin.
Copyright © 2017 Lewis et al.
Figures

References
-
- Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi:10.1056/NEJMoa1408913. - DOI - PMC - PubMed
-
- CDC 2013. Antibiotic resistance threats in the United States, 2013. CDC, Atlanta, GA.
-
- He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D’Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. doi:10.1038/ng.2478. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
