Preventing inflammation inhibits biopsy-mediated changes in tumor cell behavior
- PMID: 28790339
- PMCID: PMC5548904
- DOI: 10.1038/s41598-017-07660-4
Preventing inflammation inhibits biopsy-mediated changes in tumor cell behavior
Abstract
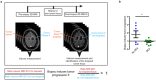
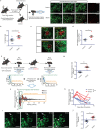
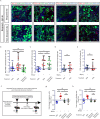
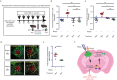
Although biopsies and tumor resection are prognostically beneficial for glioblastomas (GBM), potential negative effects have also been suggested. Here, using retrospective study of patients and intravital imaging of mice, we identify some of these negative aspects, including stimulation of proliferation and migration of non-resected tumor cells, and provide a strategy to prevent these adverse effects. By repeated high-resolution intravital microscopy, we show that biopsy-like injury in GBM induces migration and proliferation of tumor cells through chemokine (C-C motif) ligand 2 (CCL-2)-dependent recruitment of macrophages. Blocking macrophage recruitment or administrating dexamethasone, a commonly used glucocorticoid to prevent brain edema in GBM patients, suppressed the observed inflammatory response and subsequent tumor growth upon biopsy both in mice and in multifocal GBM patients. Taken together, our study suggests that inhibiting CCL-2-dependent recruitment of macrophages may further increase the clinical benefits from surgical and biopsy procedures.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Heuckmann JM, Thomas RK. A new generation of cancer genome diagnostics for routine clinical use: overcoming the roadblocks to personalized cancer medicine. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2015;26:1830–1837. doi: 10.1093/annonc/mdv184. - DOI - PubMed
-
- Tomlins SA, Palanisamy N, Siddiqui J, Chinnaiyan AM, Kunju LP. Antibody-based detection of ERG rearrangements in prostate core biopsies, including diagnostically challenging cases: ERG staining in prostate core biopsies. Archives of pathology & laboratory medicine. 2012;136:935–946. doi: 10.5858/arpa.2011-0424-OA. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

