Multi-Kernel Learning with Dartel Improves Combined MRI-PET Classification of Alzheimer's Disease in AIBL Data: Group and Individual Analyses
- PMID: 28790908
- PMCID: PMC5524673
- DOI: 10.3389/fnhum.2017.00380
Multi-Kernel Learning with Dartel Improves Combined MRI-PET Classification of Alzheimer's Disease in AIBL Data: Group and Individual Analyses
Erratum in
-
Corrigendum: Multi-Kernel Learning with Dartel Improves Combined MRI-PET Classification of Alzheimer's Disease in AIBL Data: Group and Individual Analyses.Front Hum Neurosci. 2017 Sep 8;11:457. doi: 10.3389/fnhum.2017.00457. eCollection 2017. Front Hum Neurosci. 2017. PMID: 28912705 Free PMC article.
Abstract
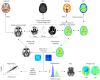
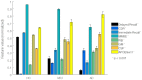
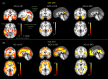
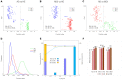
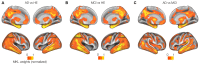
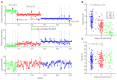
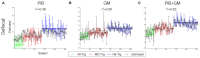
Magnetic resonance imaging (MRI) and positron emission tomography (PET) are neuroimaging modalities typically used for evaluating brain changes in Alzheimer's disease (AD). Due to their complementary nature, their combination can provide more accurate AD diagnosis or prognosis. In this work, we apply a multi-modal imaging machine-learning framework to enhance AD classification and prediction of diagnosis of subject-matched gray matter MRI and Pittsburgh compound B (PiB)-PET data related to 58 AD, 108 mild cognitive impairment (MCI) and 120 healthy elderly (HE) subjects from the Australian imaging, biomarkers and lifestyle (AIBL) dataset. Specifically, we combined a Dartel algorithm to enhance anatomical registration with multi-kernel learning (MKL) technique, yielding an average of >95% accuracy for three binary classification problems: AD-vs.-HE, MCI-vs.-HE and AD-vs.-MCI, a considerable improvement from individual modality approach. Consistent with t-contrasts, the MKL weight maps revealed known brain regions associated with AD, i.e., (para)hippocampus, posterior cingulate cortex and bilateral temporal gyrus. Importantly, MKL regression analysis provided excellent predictions of diagnosis of individuals by r2 = 0.86. In addition, we found significant correlations between the MKL classification and delayed memory recall scores with r2 = 0.62 (p < 0.01). Interestingly, outliers in the regression model for diagnosis were mainly converter samples with a higher likelihood of converting to the inclined diagnostic category. Overall, our work demonstrates the successful application of MKL with Dartel on combined neuromarkers from different neuroimaging modalities in the AIBL data. This lends further support in favor of machine learning approach in improving the diagnosis and risk prediction of AD.
Keywords: Alzheimer’s disease; Australian imaging; biomarkers; classification; lifestyle AIBL; machine learning; multi-kernel learning; prediction.
Figures

References
-
- Bergouignan L., Chupin M., Czechowska Y., Kinkingnéhun S., Lemogne C., Le Bastard G., et al. (2009). Can voxel based morphometry, manual segmentation and automated segmentation equally detect hippocampal volume differences in acute depression? Neuroimage 45, 29–37. 10.1016/j.neuroimage.2008.11.006 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

