Insight Into the Origin and Evolution of the Vibrio parahaemolyticus Pandemic Strain
- PMID: 28790994
- PMCID: PMC5523582
- DOI: 10.3389/fmicb.2017.01397
Insight Into the Origin and Evolution of the Vibrio parahaemolyticus Pandemic Strain
Abstract
A strain of Vibrio parahaemolyticus that emerged in 1995 caused the first known pandemic involving this species. This strain comprises clonal autochthonous ocean-dwelling bacteria whose evolution has occurred in the ocean environment. The low sequence diversity in this population enabled the discovery of information on its origin and evolution that has been hidden in bacterial clones that have evolved over a long period. Multilocus sequencing and microarray analysis, together with phylogenetic analysis, of pandemic and pre-pandemic isolates has suggested that the founder clone was an O3:K6 non-pathogenic strain that initially acquired a toxRS/new region and subsequently acquired at least seven novel genomic islands. Sequencing and comparison of whole genomes later confirmed these early observations, and it confirmed that most of the genetic changes occurred via gene conversion involving horizontally transmitted DNA. The highly clonal population rapidly diversified, especially in terms of antigenicity, and 27 serotypes have already been reported. Comparisons of the core genomes derived from the founder clone indicate that there are only a few hundred single-nucleotide variations between isolates. However, when the whole genome is considered (the core plus non-core genome and from any clonal frame), the amount of DNA with a different clonal frame can reach up to 4.2% and the number of single-nucleotide variations can reach several hundred thousand. Altogether, these and previous observations based on multilocus sequence typing, microarray analysis, and whole-genome sequencing indicate the large contribution made by DNA with different clonal genealogy to genome diversification. The evidence also indicates that horizontal gene transfer (HGT) caused the emergence of new pathogens. Furthermore, the extent of HGT seems to depend on the vicissitudes of the life of each bacterium, as exemplified by differences in thousands of base pairs acquired by HGT among almost identical genetic isolates.
Keywords: diarrhea; ecology; genomic; horizontal gene transfer; phylogeny.
Figures
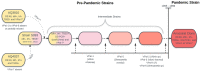
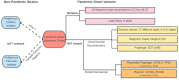
References
-
- Bhuiyan N. A., Ansaruzzaman M., Kamruzzaman M., Alam K., Chowdhury N. R., Nishibuchi M., et al. (2002). Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 40 284–286. 10.1128/JCM.40.1.284-286.2002 - DOI - PMC - PubMed
-
- Chao G., Jiao X., Zhou X., Wang F., Yang Z., Huang J., et al. (2010). Distribution of genes encoding four pathogenicity islands (VPaIs), T6SS, biofilm, and type I pilus in food and clinical strains of Vibrio parahaemolyticus in China. Foodborne Pathog. Dis. 7 649–658. 10.1089/fpd.2009.0441 - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

