Investigation of sphingosine kinase 1 in interferon responses during dengue virus infection
- PMID: 28791126
- PMCID: PMC5539417
- DOI: 10.1038/cti.2017.32
Investigation of sphingosine kinase 1 in interferon responses during dengue virus infection
Abstract
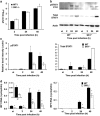
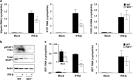
Dengue virus (DENV) regulates sphingosine kinase (SK)-1 activity and chemical inhibition of SK1 reduces DENV infection. In primary murine embryonic fibroblasts (pMEFs) lacking SK1 however, DENV infection is enhanced and this is associated with induction of normal levels of interferon beta (IFN-β) but reduced levels of IFN-stimulated genes (ISGs). We have further investigated this link between SK1 and type I IFN responses. DENV infection downregulates cell-surface IFN-alpha receptor (IFNAR)1 in both wild-type (WT) and SK1-/- pMEF, but, consistent with poor ISG responses, shows reduced induction of phosphorylated (p)-STAT1 and key IFN regulatory factors (IRF)1 and -7 in SK1-/- pMEF. Direct IFN stimulation induced ISGs (viperin, IFIT1), CXCL10, IRF1 and -7 and p-STAT1. Responses, however, were significantly reduced in SK1-/- pMEF, except for IFN-stimulated CXCL10 and IRF7. Poor IFN responses in SK1-/- pMEF were associated with a small reduction in basal cell-surface IFNAR1 and IRF1 mRNA in uninfected SK1-/- compared with WT pMEF. In contrast, treatment of cells with the SK1 inhibitor, SK1-I or expression of an inhibitory SK1 short hairpin RNA (shRNA), both of which reduce DENV infection, does not alter basal IRF1 mRNA or affect type I IFN stimulation of p-STAT1. Thus, cells genetically lacking SK1 can induce many responses normally following DENV infection, but have adaptive changes in IFNAR1 and IRF1 that compromise DENV-induced type I IFN responses. This suggests a biological link between SK1 and IFN-stimulated pathways. Other approaches to reduce SK1 activity, however, do not influence these important antiviral pathways but reduce infection and may be useful antiviral strategies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol 2008; 62: 71–92. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
