m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways
- PMID: 28792938
- PMCID: PMC5729908
- DOI: 10.1038/nature23450
m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways
Abstract
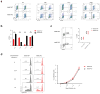
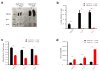
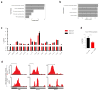
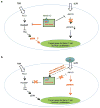
N6-methyladenosine (m6A) is the most common and abundant messenger RNA modification, modulated by 'writers', 'erasers' and 'readers' of this mark. In vitro data have shown that m6A influences all fundamental aspects of mRNA metabolism, mainly mRNA stability, to determine stem cell fates. However, its in vivo physiological function in mammals and adult mammalian cells is still unknown. Here we show that the deletion of m6A 'writer' protein METTL3 in mouse T cells disrupts T cell homeostasis and differentiation. In a lymphopaenic mouse adoptive transfer model, naive Mettl3-deficient T cells failed to undergo homeostatic expansion and remained in the naive state for up to 12 weeks, thereby preventing colitis. Consistent with these observations, the mRNAs of SOCS family genes encoding the STAT signalling inhibitory proteins SOCS1, SOCS3 and CISH were marked by m6A, exhibited slower mRNA decay and showed increased mRNAs and levels of protein expression in Mettl3-deficient naive T cells. This increased SOCS family activity consequently inhibited IL-7-mediated STAT5 activation and T cell homeostatic proliferation and differentiation. We also found that m6A has important roles for inducible degradation of Socs mRNAs in response to IL-7 signalling in order to reprogram naive T cells for proliferation and differentiation. Our study elucidates for the first time, to our knowledge, the in vivo biological role of m6A modification in T-cell-mediated pathogenesis and reveals a novel mechanism of T cell homeostasis and signal-dependent induction of mRNA degradation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures







Comment in
-
Writer's block: preventing m6A mRNA methylation promotes T cell naivety.Immunol Cell Biol. 2017 Nov;95(10):855-856. doi: 10.1038/icb.2017.67. Epub 2017 Sep 18. Immunol Cell Biol. 2017. PMID: 28925383 No abstract available.
Similar articles
-
IL-7/STAT5 cytokine signaling pathway is essential but insufficient for maintenance of naive CD4 T cell survival in peripheral lymphoid organs.J Immunol. 2007 Jan 1;178(1):262-70. doi: 10.4049/jimmunol.178.1.262. J Immunol. 2007. PMID: 17182563
-
Suppressor of cytokine signaling 1 stringently regulates distinct functions of IL-7 and IL-15 in vivo during T lymphocyte development and homeostasis.J Immunol. 2006 Apr 1;176(7):4029-41. doi: 10.4049/jimmunol.176.7.4029. J Immunol. 2006. PMID: 16547238
-
METTL3 Is Involved in the Development of Graves' Disease by Inducing SOCS mRNA m6A Modification.Front Endocrinol (Lausanne). 2021 Sep 20;12:666393. doi: 10.3389/fendo.2021.666393. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34616359 Free PMC article.
-
The dynamic epitranscriptome: N6-methyladenosine and gene expression control.Nat Rev Mol Cell Biol. 2014 May;15(5):313-26. doi: 10.1038/nrm3785. Epub 2014 Apr 9. Nat Rev Mol Cell Biol. 2014. PMID: 24713629 Free PMC article. Review.
-
SOCS proteins: negative regulators of cytokine signaling.Stem Cells. 2001;19(5):378-87. doi: 10.1634/stemcells.19-5-378. Stem Cells. 2001. PMID: 11553846 Review.
Cited by
-
Role and regulators of N6-methyladenosine (m6A) RNA methylation in inflammatory subtypes of asthma: a comprehensive review.Front Pharmacol. 2024 Jul 23;15:1360607. doi: 10.3389/fphar.2024.1360607. eCollection 2024. Front Pharmacol. 2024. PMID: 39108751 Free PMC article. Review.
-
The RNA m6A writer METTL3 in tumor microenvironment: emerging roles and therapeutic implications.Front Immunol. 2024 Jan 22;15:1335774. doi: 10.3389/fimmu.2024.1335774. eCollection 2024. Front Immunol. 2024. PMID: 38322265 Free PMC article. Review.
-
A New Model of Spontaneous Colitis in Mice Induced by Deletion of an RNA m6A Methyltransferase Component METTL14 in T Cells.Cell Mol Gastroenterol Hepatol. 2020;10(4):747-761. doi: 10.1016/j.jcmgh.2020.07.001. Epub 2020 Jul 4. Cell Mol Gastroenterol Hepatol. 2020. PMID: 32634481 Free PMC article.
-
IL-7: A promising adjuvant ensuring effective T cell responses and memory in combination with cancer vaccines?Front Immunol. 2022 Oct 28;13:1022808. doi: 10.3389/fimmu.2022.1022808. eCollection 2022. Front Immunol. 2022. PMID: 36389666 Free PMC article. Review.
-
Regulation of N6-Methyladenosine in the Differentiation of Cancer Stem Cells and Their Fate.Front Cell Dev Biol. 2020 Sep 22;8:561703. doi: 10.3389/fcell.2020.561703. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33072746 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

