Investigation of the Melting Behavior of DNA Three-Way Junctions in the Closed and Open States
- PMID: 28793208
- PMCID: PMC5549643
- DOI: 10.1016/j.bpj.2017.06.024
Investigation of the Melting Behavior of DNA Three-Way Junctions in the Closed and Open States
Abstract
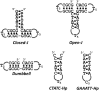
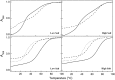
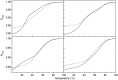
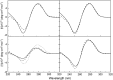
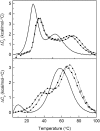
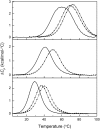
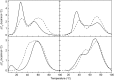
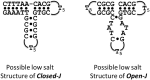
Intramolecular three-way junctions are commonly found in both DNA and RNA. These structures are functionally relevant in ribozymes, riboswitches, rRNA, and during replication. In this work, we present a thermodynamic description of the unfolding of DNA intramolecular three-way junctions. We used a combination of spectroscopic and calorimetric techniques to investigate the folding/unfolding thermodynamics of two three-way junctions with a closed (Closed-J) or open (Open-J) junction and their appropriate control stem-loop motifs (GAAATT-Hp, CTATC-Hp, and Dumbbell). The overall results show that both junctions are stable over a wide range of salt concentrations. However, Open-J is more stable due to a higher enthalpy contribution from the formation of a higher number of basepair stacks whereas Closed-J has a defined structure and retains the basepair stacking of all three stems. The comparison of the experimental results of Closed-J and Open-J with those of their component stem-loop motifs allowed us to be more specific about their cooperative unfolding. For instance, Closed-J sacrifices thermal stability of the Dumbbell structure to maintain an overall folded state. At higher salt concentration, the simultaneous unfolding of the above domains is lost, resulting in the unfolding of the three separate stems. In contrast, the junction of Open-J in low salt retains the thermal and enthalpic stability of the Dumbbell structure although sacrificing stability of the CTATC stem. The relative stability of Dumbbell is the primary reason for the higher ΔG°(5), or free energy, value seen for Open-J at low salt. Higher salt not only maintains thermal stability of the Dumbbell structure in Open-J but causes the CTATC stem to fully fold.
Copyright © 2017 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Batey R.T., Gilbert S.D., Montange R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

