Iron affects Ire1 clustering propensity and the amplitude of endoplasmic reticulum stress signaling
- PMID: 28794014
- PMCID: PMC5665437
- DOI: 10.1242/jcs.201715
Iron affects Ire1 clustering propensity and the amplitude of endoplasmic reticulum stress signaling
Abstract
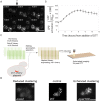
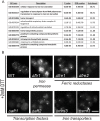
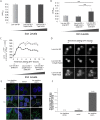
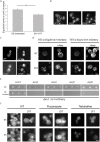
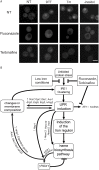
The unfolded protein response (UPR) allows cells to adjust secretory pathway capacity according to need. Ire1, the endoplasmic reticulum (ER) stress sensor and central activator of the UPR is conserved from the budding yeast Saccharomyces cerevisiae to humans. Under ER stress conditions, Ire1 clusters into foci that enable optimal UPR activation. To discover factors that affect Ire1 clustering, we performed a high-content screen using a whole-genome yeast mutant library expressing Ire1-mCherry. We imaged the strains following UPR induction and found 154 strains that displayed alterations in Ire1 clustering. The hits were enriched for iron and heme effectors and binding proteins. By performing pharmacological depletion and repletion, we confirmed that iron (Fe3+) affects UPR activation in both yeast and human cells. We suggest that Ire1 clustering propensity depends on membrane composition, which is governed by heme-dependent biosynthesis of sterols. Our findings highlight the diverse cellular functions that feed into the UPR and emphasize the cross-talk between organelles required to concertedly maintain homeostasis.
Keywords: Heme; Ire1; Iron; Saccharomyces cerevisiae; Sterol; UPR.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
-
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P. and Boeke J.D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2 - DOI - PubMed
-
- Breslow D. K., Cameron D. M., Collins S. R., Schuldiner M., Stewart-Ornstein J., Newman H. W., Braun S., Madhani H. D., Krogan N. J. and Weissman J. S. (2008). A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5, 711-718. 10.1038/nmeth.1234 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

