Reverse Genetics System Demonstrates that Rotavirus Nonstructural Protein NSP6 Is Not Essential for Viral Replication in Cell Culture
- PMID: 28794037
- PMCID: PMC5640853
- DOI: 10.1128/JVI.00695-17
Reverse Genetics System Demonstrates that Rotavirus Nonstructural Protein NSP6 Is Not Essential for Viral Replication in Cell Culture
Abstract
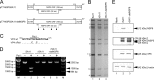
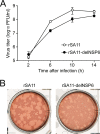
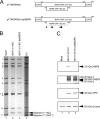
The use of overlapping open reading frames (ORFs) to synthesize more than one unique protein from a single mRNA has been described for several viruses. Segment 11 of the rotavirus genome encodes two nonstructural proteins, NSP5 and NSP6. The NSP6 ORF is present in the vast majority of rotavirus strains, and therefore the NSP6 protein would be expected to have a function in viral replication. However, there is no direct evidence of its function or requirement in the viral replication cycle yet. Here, taking advantage of a recently established plasmid-only-based reverse genetics system that allows rescue of recombinant rotaviruses entirely from cloned cDNAs, we generated NSP6-deficient viruses to directly address its significance in the viral replication cycle. Viable recombinant NSP6-deficient viruses could be engineered. Single-step growth curves and plaque formation of the NSP6-deficient viruses confirmed that NSP6 expression is of limited significance for RVA replication in cell culture, although the NSP6 protein seemed to promote efficient virus growth.IMPORTANCE Rotavirus is one of the most important pathogens of severe diarrhea in young children worldwide. The rotavirus genome, consisting of 11 segments of double-stranded RNA, encodes six structural proteins (VP1 to VP4, VP6, and VP7) and six nonstructural proteins (NSP1 to NSP6). Although specific functions have been ascribed to each of the 12 viral proteins, the role of NSP6 in the viral replication cycle remains unknown. In this study, we demonstrated that the NSP6 protein is not essential for viral replication in cell culture by using a recently developed plasmid-only-based reverse genetics system. This reverse genetics approach will be successfully applied to answer questions of great interest regarding the roles of rotaviral proteins in replication and pathogenicity, which can hardly be addressed by conventional approaches.
Keywords: NSP6; reverse genetics; rotavirus; viral replication.
Copyright © 2017 American Society for Microbiology.
Figures


References
-
- Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, World Health Organization-Coordinated Global Rotavirus Surveillance Network. 2016. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000-2013. Clin Infect Dis 62(Suppl 2):S96–S105. doi: 10.1093/cid/civ1013. - DOI - PMC - PubMed
-
- Estes MK, Greenberg HB. 2013. Rotaviruses, p 1347–1401. In Knipe DM, Howley PM (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

