Molecular identification of Nocardia species using the sod A gene: Identificación molecular de especies de Nocardia utilizando el gen sodA
- PMID: 28794885
- PMCID: PMC5547243
- DOI: 10.1016/j.nmni.2017.03.008
Molecular identification of Nocardia species using the sod A gene: Identificación molecular de especies de Nocardia utilizando el gen sodA
Abstract
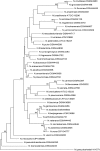
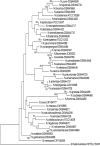
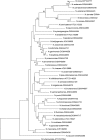
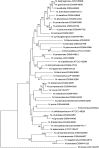
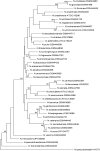
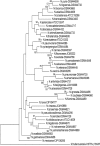
Currently for bacterial identification and classification the rrs gene encoding 16S rRNA is used as a reference method for the analysis of strains of the genus Nocardia. However, it does not have enough polymorphism to differentiate them at the species level. This fact makes it necessary to search for molecular targets that can provide better identification. The sodA gene (encoding the enzyme superoxide dismutase) has had good results in identifying species of other Actinomycetes. In this study the sodA gene is proposed for the identification and differentiation at the species level of the genus Nocardia. We used 41 type species of various collections; a 386 bp fragment of the sodA gene was amplified and sequenced, and a phylogenetic analysis was performed comparing the genes rrs (1171 bp), hsp65 (401 bp), secA1 (494 bp), gyrB (1195 bp) and rpoB (401 bp). The sequences were aligned using the Clustal X program. Evolutionary trees according to the neighbour-joining method were created with the programs Phylo_win and MEGA 6. The specific variability of the sodA genus of the genus Nocardia was analysed. A high phylogenetic resolution, significant genetic variability, and specificity and reliability were observed for the differentiation of the isolates at the species level. The polymorphism observed in the sodA gene sequence contains variable regions that allow the discrimination of closely related Nocardia species. The clear specificity, despite its small size, proves to be of great advantage for use in taxonomic studies and clinical diagnosis of the genus Nocardia.
Actualmente, para la identificación y clasificación bacteriana se utiliza como método de referencia la secuenciación el gen rrs que codifica al rRNA16S, en el caso del análisis de cepas del género Nocardia, sin embargo, no tiene el suficiente polimorfismo para diferenciarlas a nivel de especie lo que hace necesaria la búsqueda de blancos moleculares que puedan proporcionar una mejor identificación. El gen sodA (que codifica la enzima superóxido dismutasa) ha tenido buenos resultados en la identificación de especies de otros Actinomicetos. En este estudio se propone para la identificación y diferenciación a nivel de especie del género Nocardia. Se utilizaron 41 especies Tipo de diversas colecciones, se amplificó y secuenció un fragmento de 386 pb del gen sodA y se realizó un análisis filogenético comparando los genes rrs (1171 pb) hsp65(401pb) secA1 (494pb), gyrB (1195pb) y rpoB (401pb), las secuencias fueron alineadas utilizando el programa Clustal X, los árboles evolutivos de acuerdo con el método de “Neighbor-Joining”se hicieron con el programa Phylo_win y Mega 6. Se analizó la variabilidad específica del gen sodA del género Nocardia presentando una alta resolución filogenética, una variabilidad genética importante, especificidad y confiabilidad para la diferenciación de los aislados a nivel de especie. El polimorfismo observado en la secuencia del gen sodA contiene regiones variables que posibilitan la discriminación de especies de Nocardia estrechamente relacionadas, y una clara especificidad, a pesar de su pequeño tamaño, demostrando ser de gran ventaja para utilizarse en estudios taxonómicos y en el diagnóstico clínico del género Nocardia.
Keywords: Nocardia; hsp65; polymorphism; rrs; sodA.
Figures






References
-
- Deshpande L.M., Fritsche R.N., Jones R.N. Molecular epidemiology of selected multidrug-resistant bacteria: a global report from the SENTRY Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis. 2004;4:231–236. - PubMed
-
- Yassin F.A., Rainey A.F., Burghardt J., Brzezinka H., Mauch M., Schaal P.K. Nocardia paucivorans sp. nov. Int J Syst Evol Microbiol. 2000;50:803–809. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
