Risk score based on ten lncRNA-mRNA expression predicts the survival of stage II-III colorectal carcinoma
- PMID: 28796819
- PMCID: PMC5552098
- DOI: 10.1371/journal.pone.0182908
Risk score based on ten lncRNA-mRNA expression predicts the survival of stage II-III colorectal carcinoma
Erratum in
-
Correction: Risk score based on ten lncRNA-mRNA expression predicts the survival of stage II-III colorectal carcinoma.PLoS One. 2018 Aug 2;13(8):e0201993. doi: 10.1371/journal.pone.0201993. eCollection 2018. PLoS One. 2018. PMID: 30071109 Free PMC article.
Abstract
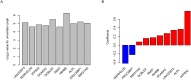
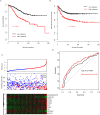
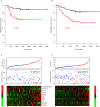
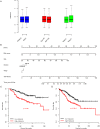
The prognosis of colorectal carcinoma (CRC) is unstable in the stage II-III patients. Patients with early stage II CRC have a relative poor prognosis while other stage III CRC patients have a better prognosis. In our work, by utilizing the expression of lncRNAs and mRNAs measured by microarray (GSE39582), we constructed a risk score staging system with Cox multivariate regression model to predict the outcome of grade II-III CRC patients. Ten genes including two lncRNAs and eight mRNAs were used to estimate the survival of stage II-III CRC patients. The patients with high risk scores have poorer survival rate those with low risk scores, significantly. These results were further validated in another three independent datasets (GSE37892, GSE33113, and GSE17536). The relationship between clinical information and were evaluated, and the risk score is independent from the other clinical information and performs better in evaluating the survival of stage II-III CRC patients. Moreover, the correlation between chemotherapy was also evaluated, and we found that both patients with or without chemotherapy have a poor survival in high risk group. Gene Set Enrichment Analysis were used to find the difference between high-risk and low-risk groups, and pathways including cell adhesion and focal adhesion were significantly enriched, suggesting that the risk score reflects the status of cell-cell physical interaction. In summary, we constructed a risk staging model for grade II-III CRC, which is independent from and performs better than clinical information.
Conflict of interest statement
Figures
Similar articles
-
Identification of key lncRNAs in colorectal cancer progression based on associated protein-protein interaction analysis.World J Surg Oncol. 2017 Aug 10;15(1):153. doi: 10.1186/s12957-017-1211-7. World J Surg Oncol. 2017. PMID: 28797257 Free PMC article.
-
Regulatory activity based risk model identifies survival of stage II and III colorectal carcinoma.Oncotarget. 2017 Sep 28;8(58):98360-98370. doi: 10.18632/oncotarget.21312. eCollection 2017 Nov 17. Oncotarget. 2017. PMID: 29228695 Free PMC article.
-
ColoGuideEx: a robust gene classifier specific for stage II colorectal cancer prognosis.Gut. 2012 Nov;61(11):1560-7. doi: 10.1136/gutjnl-2011-301179. Epub 2012 Jan 2. Gut. 2012. PMID: 22213796
-
Elevated serum level of lncRNA-HIF1A-AS1 as a novel diagnostic predictor for worse prognosis in colorectal carcinoma.Cancer Biomark. 2017 Dec 6;20(4):417-424. doi: 10.3233/CBM-170179. Cancer Biomark. 2017. PMID: 28946548
-
Clinical significance of perineural invasion in stages II and III colorectal cancer.Pathol Res Pract. 2015 Nov;211(11):839-44. doi: 10.1016/j.prp.2015.09.001. Epub 2015 Sep 4. Pathol Res Pract. 2015. PMID: 26429129
Cited by
-
Correction: Risk score based on ten lncRNA-mRNA expression predicts the survival of stage II-III colorectal carcinoma.PLoS One. 2018 Aug 2;13(8):e0201993. doi: 10.1371/journal.pone.0201993. eCollection 2018. PLoS One. 2018. PMID: 30071109 Free PMC article.
-
A 6 lncRNA-Based Risk Score System for Predicting the Recurrence of Colon Adenocarcinoma Patients.Front Oncol. 2020 Feb 6;10:81. doi: 10.3389/fonc.2020.00081. eCollection 2020. Front Oncol. 2020. PMID: 32117736 Free PMC article.
-
Identification of MYC and STAT3 for early diagnosis based on the long noncoding RNA-mRNA network and bioinformatics in colorectal cancer.Front Immunol. 2025 Jan 3;15:1497919. doi: 10.3389/fimmu.2024.1497919. eCollection 2024. Front Immunol. 2025. PMID: 39830506 Free PMC article.
References
-
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108. Epub 2015/02/06. doi: 10.3322/caac.21262 . - DOI - PubMed
-
- Siegel R, Miller K, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65(1):5–29. - PubMed
-
- Saus E, Brunet-Vega A, Iraola-Guzman S, Pegueroles C, Gabaldon T, Pericay C. Long Non-Coding RNAs As Potential Novel Prognostic Biomarkers in Colorectal Cancer. Frontiers in genetics. 2016;7:54 Epub 2016/05/06. doi: 10.3389/fgene.2016.00054 ; - DOI - PMC - PubMed
-
- Li Y, Huang S, Li Y, Zhang W, He K, Zhao M, et al. Decreased expression of LncRNA SLC25A25-AS1 promotes proliferation, chemoresistance, and EMT in colorectal cancer cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(10):14205–15. Epub 2016/08/25. doi: 10.1007/s13277-016-5254-0 . - DOI - PubMed
-
- Lian Y, Ding J, Zhang Z, Shi Y, Zhu Y, Li J, et al. The long noncoding RNA HOXA transcript at the distal tip promotes colorectal cancer growth partially via silencing of p21 expression. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(6):7431–40. Epub 2015/12/19. doi: 10.1007/s13277-015-4617-2 . - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

