ODG: Omics database generator - a tool for generating, querying, and analyzing multi-omics comparative databases to facilitate biological understanding
- PMID: 28797229
- PMCID: PMC5553995
- DOI: 10.1186/s12859-017-1777-7
ODG: Omics database generator - a tool for generating, querying, and analyzing multi-omics comparative databases to facilitate biological understanding
Abstract
Background: Rapid generation of omics data in recent years have resulted in vast amounts of disconnected datasets without systemic integration and knowledge building, while individual groups have made customized, annotated datasets available on the web with few ways to link them to in-lab datasets. With so many research groups generating their own data, the ability to relate it to the larger genomic and comparative genomic context is becoming increasingly crucial to make full use of the data.
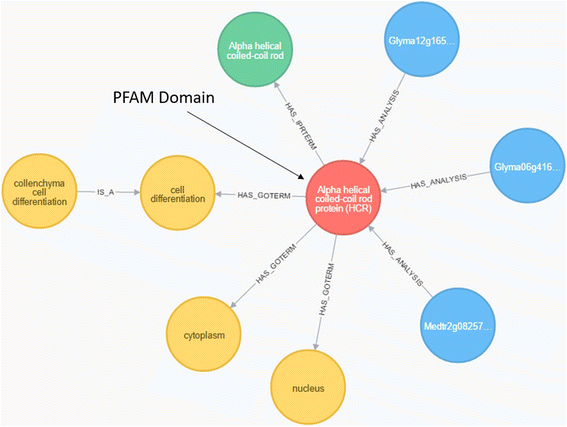
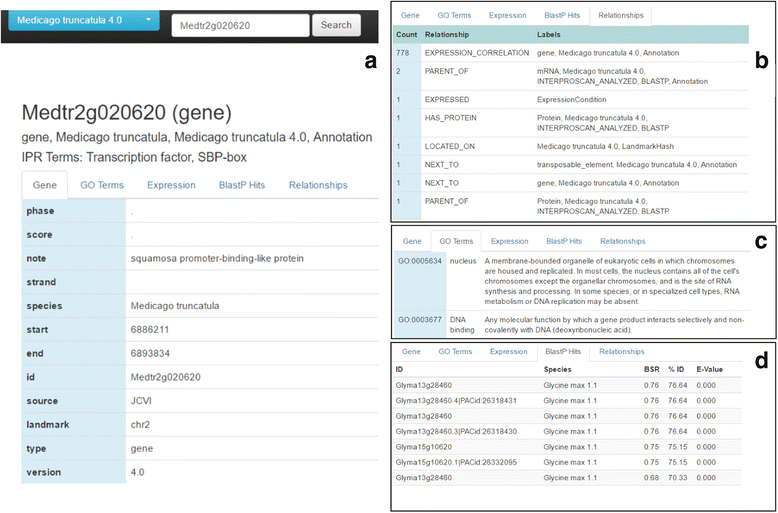
Results: The Omics Database Generator (ODG) allows users to create customized databases that utilize published genomics data integrated with experimental data which can be queried using a flexible graph database. When provided with omics and experimental data, ODG will create a comparative, multi-dimensional graph database. ODG can import definitions and annotations from other sources such as InterProScan, the Gene Ontology, ENZYME, UniPathway, and others. This annotation data can be especially useful for studying new or understudied species for which transcripts have only been predicted, and rapidly give additional layers of annotation to predicted genes. In better studied species, ODG can perform syntenic annotation translations or rapidly identify characteristics of a set of genes or nucleotide locations, such as hits from an association study. ODG provides a web-based user-interface for configuring the data import and for querying the database. Queries can also be run from the command-line and the database can be queried directly through programming language hooks available for most languages. ODG supports most common genomic formats as well as generic, easy to use tab-separated value format for user-provided annotations.
Conclusions: ODG is a user-friendly database generation and query tool that adapts to the supplied data to produce a comparative genomic database or multi-layered annotation database. ODG provides rapid comparative genomic annotation and is therefore particularly useful for non-model or understudied species. For species for which more data are available, ODG can be used to conduct complex multi-omics, pattern-matching queries.
Keywords: Annotation; Comparative genomics; Data integration; Graph database; Non-model species.
Conflict of interest statement
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Neo4j: The World’s Leading Graph Database. http://neo4j.com/. Accessed 10 Mar 2017.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

