Nutrient supply affects the mRNA expression profile of the porcine skeletal muscle
- PMID: 28797239
- PMCID: PMC5553784
- DOI: 10.1186/s12864-017-3986-x
Nutrient supply affects the mRNA expression profile of the porcine skeletal muscle
Abstract
Background: The genetic basis of muscle fat deposition in pigs is not well known. So far, we have only identified a limited number of genes involved in the absorption, transport, storage and catabolism of lipids. Such information is crucial to interpret, from a biological perspective, the results of genome-wide association analyses for intramuscular fat content and composition traits. Herewith, we have investigated how the ingestion of food changes gene expression in the gluteus medius muscle of Duroc pigs.
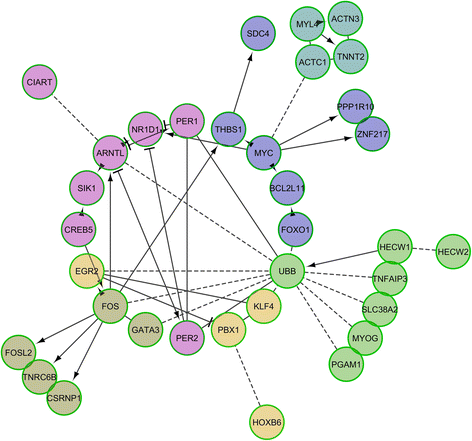
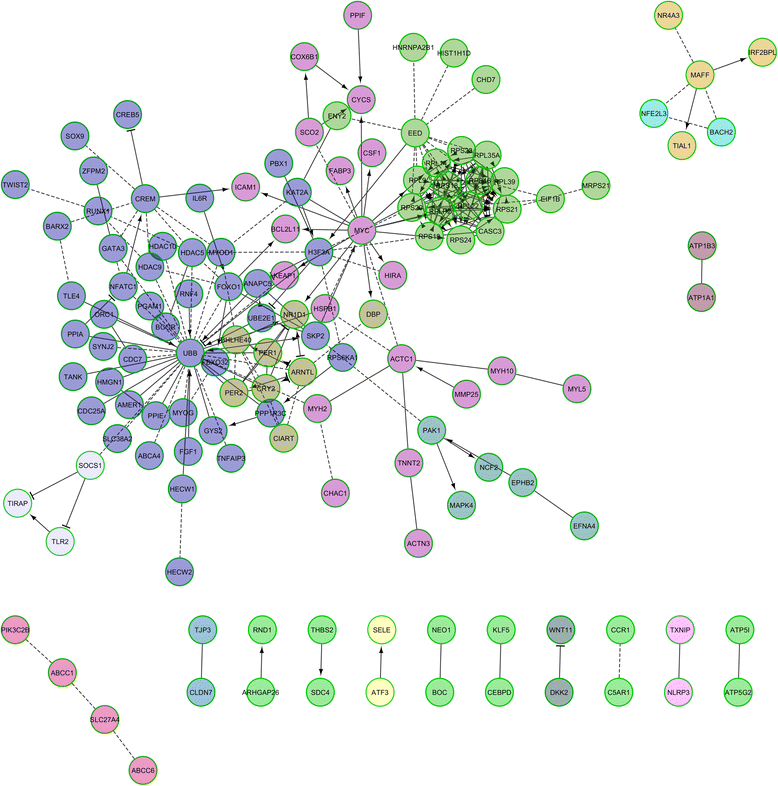
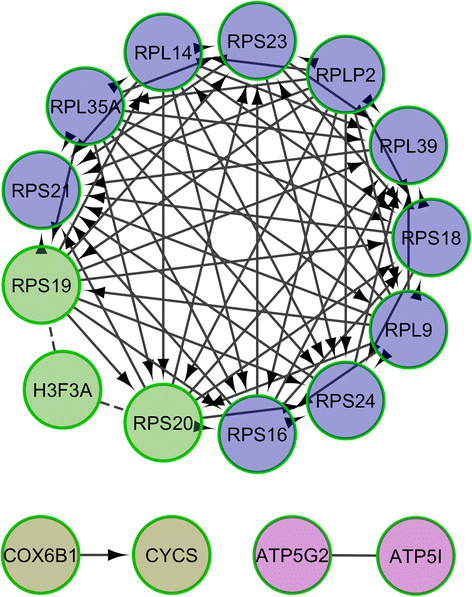
Results: By comparing the muscle mRNA expression of fasted pigs (T0) with that of pigs sampled 5 h (T1) and 7 h (T2) after food intake, we have detected differential expression (DE) for 148 (T0-T1), 520 (T0-T2) and 135 (T1-T2) genes (q-value <0.05 and a |FC| > of 1.5). Many of these DE genes were transcription factors, suggesting that we have detected the coordinated response of the skeletal muscle to nutrient supply. We also found DE genes with a dual role in oxidative stress and angiogenesis (THBS1, THBS2 and TXNIP), two biological processes that are probably activated in the post-prandial state. Finally, we have identified several loci playing a key role in the modulation of circadian rhythms (ARNTL, PER1, PER2, BHLHE40, NR1D1, SIK1, CIART and CRY2), a result that indicates that the porcine muscle circadian clock is modulated by nutrition.
Conclusion: We have shown that hundreds of genes change their expression in the porcine skeletal muscle in response to nutrient intake. Many of these loci do not have a known metabolic role, a result that suggests that our knowledge about the genetic basis of muscle energy homeostasis is still incomplete.
Keywords: Angiogenesis; Circadian rhythm; Oxidative stress; Pig; RNA-seq; Transcription factor.
Conflict of interest statement
Ethics approval
Animal care, management procedures and blood sampling were performed following national guidelines for the Good Experimental Practices and they were approved by the Ethical Committee of the Institut de Recerca i Tecnologia Agroalimentàries (IRTA).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Ayuso M, Fernández A, Núñez Y, Benítez R, Isabel B, Barragán C, et al. Comparative analysis of muscle transcriptome between pig genotypes identifies genes and regulatory mechanisms associated to growth, fatness and metabolism. PLoS One. 2015;10:e0145162. doi: 10.1371/journal.pone.0145162. - DOI - PMC - PubMed
-
- Khetarpal SA, Rader DJ. Genetics of lipid traits: genome-wide approaches yield new biology and clues to causality in coronary artery disease. Biochim Biophys Acta. 1842;2014:2010–2020. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

