LungMAP: The Molecular Atlas of Lung Development Program
- PMID: 28798251
- PMCID: PMC5792185
- DOI: 10.1152/ajplung.00139.2017
LungMAP: The Molecular Atlas of Lung Development Program
Abstract
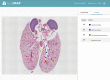
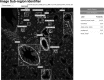
The National Heart, Lung, and Blood Institute is funding an effort to create a molecular atlas of the developing lung (LungMAP) to serve as a research resource and public education tool. The lung is a complex organ with lengthy development time driven by interactive gene networks and dynamic cross talk among multiple cell types to control and coordinate lineage specification, cell proliferation, differentiation, migration, morphogenesis, and injury repair. A better understanding of the processes that regulate lung development, particularly alveologenesis, will have a significant impact on survival rates for premature infants born with incomplete lung development and will facilitate lung injury repair and regeneration in adults. A consortium of four research centers, a data coordinating center, and a human tissue repository provides high-quality molecular data of developing human and mouse lungs. LungMAP includes mouse and human data for cross correlation of developmental processes across species. LungMAP is generating foundational data and analysis, creating a web portal for presentation of results and public sharing of data sets, establishing a repository of young human lung tissues obtained through organ donor organizations, and developing a comprehensive lung ontology that incorporates the latest findings of the consortium. The LungMAP website (www.lungmap.net) currently contains more than 6,000 high-resolution lung images and transcriptomic, proteomic, and lipidomic human and mouse data and provides scientific information to stimulate interest in research careers for young audiences. This paper presents a brief description of research conducted by the consortium, database, and portal development and upcoming features that will enhance the LungMAP experience for a community of users.
Keywords: 3D imaging; lung development; lung imaging; lung omics; single cell analysis; web resource.
Copyright © 2017 the American Physiological Society.
Figures




References
-
- Clair G, Piehowski PD, Nicola T, Kitzmiller JA, Huang EL, Zink EM, Sontag RL, Orton DJ, Moore RJ, Carson JP, Smith RD, Whitsett JA, Corley RA, Ambalavanan N, Ansong C. Spatially-resolved proteomics: rapid quantitative analysis of laser capture microdissected alveolar tissue samples. Sci Rep 6: 39223, 2016. doi:10.1038/srep39223. - DOI - PMC - PubMed
-
- Dautel SE, Kyle JE, Clair G, Sontag RL, Weitz KK, Shukla AK, Nguyen SN, Kim YM, Zink EM, Luders T, Frevert CW, Gharib SA, Laskin J, Carson JP, Metz TO, Corley RA, Ansong C. Lipidomics reveals dramatic lipid compositional changes in the maturing postnatal lung. Sci Rep 7: 40555, 2017. doi:10.1038/srep40555. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- U01 HL148861/HL/NHLBI NIH HHS/United States
- R01 HL136722/HL/NHLBI NIH HHS/United States
- UL1 TR002001/TR/NCATS NIH HHS/United States
- UL1 TR000042/TR/NCATS NIH HHS/United States
- U01 HL122681/HL/NHLBI NIH HHS/United States
- U01 HL122703/HL/NHLBI NIH HHS/United States
- U01 HL122626/HL/NHLBI NIH HHS/United States
- R01 HL095580/HL/NHLBI NIH HHS/United States
- U01 HL122642/HL/NHLBI NIH HHS/United States
- U01 HL122700/HL/NHLBI NIH HHS/United States
- P30 CA013148/CA/NCI NIH HHS/United States
- U01 HL122638/HL/NHLBI NIH HHS/United States
- U01 HL134745/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

