Metabolic networks for nitrogen utilization in Prevotella ruminicola 23
- PMID: 28798330
- PMCID: PMC5552732
- DOI: 10.1038/s41598-017-08463-3
Metabolic networks for nitrogen utilization in Prevotella ruminicola 23
Abstract
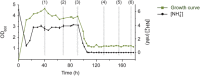
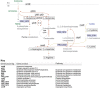
Nitrogen metabolism in gut systems remains poorly studied in spite of its importance for microbial growth and its implications for the metabolism of the host. Prevotella spp. are the most predominant bacteria detected in the rumen, but their presence has also been related to health and disease states in the human gut and oral cavity. To explore the metabolic networks for nitrogen assimilation in this bacterium, changes in gene expression profiles in response to variations in the available nitrogen source and to different concentrations of ammonium were analyzed by microarray and reverse transcription quantitative PCR, and linked with function by further proteomic analysis. The observed patterns of transcript abundances for genes involved in ammonium assimilation differed from the classical "enteric paradigm" for nitrogen utilization. Expression of genes encoding high substrate affinity nitrogen assimilation enzymes (GS-GOGAT system) was similar in growth-limiting and non-limiting nitrogen concentrations in P. ruminicola 23, whereas E. coli and Salmonella spp. responses to excess nitrogen involve only low substrate affinity enzymes. This versatile behavior might be a key feature for ecological success in habitats such as the rumen and human colon where nitrogen is rarely limiting for growth, and might be linked to previously reported Prevotella spp. population imbalances relative to other bacterial species in gut systems.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Gasparic A, et al. Isolation of genes encoding β-D-xylanase, β-D-xylosidase and α-L-arabinofuranosidase activities from the rumen bacterium Prevotella ruminicola B14. FEMS Microbiol. Lett. 1995;125:135–141. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

