Characterization of the Culturable Subpopulations of Lactobacillus in the Chicken Intestinal Tract as a Resource for Probiotic Development
- PMID: 28798730
- PMCID: PMC5526839
- DOI: 10.3389/fmicb.2017.01389
Characterization of the Culturable Subpopulations of Lactobacillus in the Chicken Intestinal Tract as a Resource for Probiotic Development
Abstract
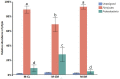
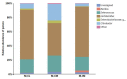
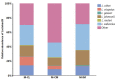
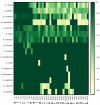
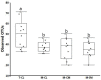
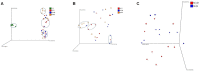
To gain better understanding of the distributions of the culturable Lactobacillus species in the chicken intestinal tract, we collected ceca, and distal ileum from 10 3-weeks-old broiler chickens. Lactobacillus strains from cecal lumen contents (M-CL), and those associated with mucosa of ceca (M-CM) and ileum (M-IM) were recovered on de Man, Rogosa and Sharpe (MRS) agar plates, and used for microbiota analysis. The total cecal content (T-CL) was also used directly for microbiota analysis. We purposefully focused on MRS-recovered populations to gain understanding of the culturable subpopulations of Lactobacillus, since the culturability is an important phenotype in order to exploit the chicken gut microbiota as a resource for development of probiotics. The V1-V3 regions of 16S rRNA gene was amplified from genomic DNA samples, and the pooled amplicons were analyzed by MiSeq sequencing with paired-end read 300 cycle option. Among MRS groups, Firmicutes were significantly higher in M-IM and M-CL as compared to M-CM, whereas Proteobacteria were significantly higher in M-CM as compared to M-IM and M-CL at p < 0.05. Among Lactobacillus, L. salivarius (36%) and L. johnsonii (21%) were higher in M-IM as compared to M-CL (L. salivarius, 28%; L. johnsonii, 15%), and M-CM (L. salivarius, 20%; L. johnsonii, 11%). L. crispatus was found significantly higher in M-CL as compared to M-IM (p < 0.01) whereas L. gasseri was found significantly higher in M-IM as compared to M-CM (p < 0.05). L. aviarius, and L. fornicalis were only observed in T-CL. In summary, Lactobacillus populations recovered on MRS vary with different regions and locations in chicken GIT, which might indicate their distinct functional roles in different gastrointestinal tract (GIT) niches, and some species of Lactobacillus are not culturable on MRS agar media. This study is the first attempt to define culturable Lactobacillus subpopulations in the chicken intestinal tract comprehensively using 16S rRNA gene profiling, and the findings of this study will be used as a platform to develop a new strategy for isolation of effective Lactobacillus probiotic candidates based on comparative analyses of chicken gut microbiota.
Keywords: Lactobacillus; broiler; gastrointestinal tract; microbiota; probiotics.
Figures
References
-
- Ahasan A., Agazzi A., Invernizzi G., Bontempo V., Savoini G. (2015). The beneficial role of probiotics in monogastric animal nutrition and health. J. Dairy Vet. Anim. Res. 2:00041 10.15406/jdvar.2015.02.00041 - DOI
-
- Anderson R. C., Cookson A. L., McNabb W. C., Park Z., McCann M. J., Kelly W. J., et al. (2010). Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol. 10:316 10.1186/1471-2180-10-316 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

