Method to Assemble Genomic DNA Fragments or Genes on Human Artificial Chromosome with Regulated Kinetochore Using a Multi-Integrase System
- PMID: 28799737
- PMCID: PMC5778389
- DOI: 10.1021/acssynbio.7b00209
Method to Assemble Genomic DNA Fragments or Genes on Human Artificial Chromosome with Regulated Kinetochore Using a Multi-Integrase System
Abstract
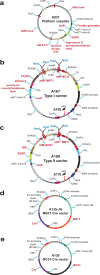
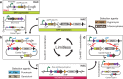
The production of cells capable of carrying multiple transgenes to Mb-size genomic loci has multiple applications in biomedicine and biotechnology. In order to achieve this goal, three key steps are required: (i) cloning of large genomic segments; (ii) insertion of multiple DNA blocks at a precise location and (iii) the capability to eliminate the assembled region from cells. In this study, we designed the iterative integration system (IIS) that utilizes recombinases Cre, ΦC31 and ΦBT1, and combined it with a human artificial chromosome (HAC) possessing a regulated kinetochore (alphoidtetO-HAC). We have demonstrated that the IIS-alphoidtetO-HAC system is a valuable genetic tool by reassembling a functional gene from multiple segments on the HAC. IIS-alphoidtetO-HAC has several notable advantages over other artificial chromosome-based systems. This includes the potential to assemble an unlimited number of genomic DNA segments; a DNA assembly process that leaves only a small insertion (<60 bp) scar between adjacent DNA, allowing genes reassembled from segments to be spliced correctly; a marker exchange system that also changes cell color, and counter-selection markers at each DNA insertion step, simplifying selection of correct clones; and presence of an error proofing mechanism to remove cells with misincorporated DNA segments, which improves the integrity of assembly. In addition, the IIS-alphoidtetO-HAC carrying a locus of interest is removable, offering the unique possibility to revert the cell line to its pretransformed state and compare the phenotypes of human cells with and without a functional copy of a gene(s). Thus, IIS-alphoidtetO-HAC allows investigation of complex biomedical pathways, gene(s) regulation, and has the potential to engineer synthetic chromosomes with a predetermined set of genes.
Keywords: DNA assembly; HAC; IIS; human artificial chromosome; iterative integration system; synthetic biology.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Oshimura M., Kazuki Y., Iida Y., and Uno N. (2013) New Vectors for Gene Delivery: Human and Mouse Artificial Chromosomes, In eLS, John Wiley & Sons, Ltd..
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

