The nucleoid protein Dps binds genomic DNA of Escherichia coli in a non-random manner
- PMID: 28800583
- PMCID: PMC5553809
- DOI: 10.1371/journal.pone.0182800
The nucleoid protein Dps binds genomic DNA of Escherichia coli in a non-random manner
Abstract
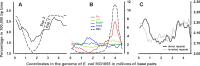
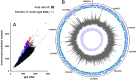
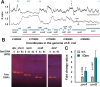
Dps is a multifunctional homododecameric protein that oxidizes Fe2+ ions accumulating them in the form of Fe2O3 within its protein cavity, interacts with DNA tightly condensing bacterial nucleoid upon starvation and performs some other functions. During the last two decades from discovery of this protein, its ferroxidase activity became rather well studied, but the mechanism of Dps interaction with DNA still remains enigmatic. The crucial role of lysine residues in the unstructured N-terminal tails led to the conventional point of view that Dps binds DNA without sequence or structural specificity. However, deletion of dps changed the profile of proteins in starved cells, SELEX screen revealed genomic regions preferentially bound in vitro and certain affinity of Dps for artificial branched molecules was detected by atomic force microscopy. Here we report a non-random distribution of Dps binding sites across the bacterial chromosome in exponentially growing cells and show their enrichment with inverted repeats prone to form secondary structures. We found that the Dps-bound regions overlap with sites occupied by other nucleoid proteins, and contain overrepresented motifs typical for their consensus sequences. Of the two types of genomic domains with extensive protein occupancy, which can be highly expressed or transcriptionally silent only those that are enriched with RNA polymerase molecules were preferentially occupied by Dps. In the dps-null mutant we, therefore, observed a differentially altered expression of several targeted genes and found suppressed transcription from the dps promoter. In most cases this can be explained by the relieved interference with Dps for nucleoid proteins exploiting sequence-specific modes of DNA binding. Thus, protecting bacterial cells from different stresses during exponential growth, Dps can modulate transcriptional integrity of the bacterial chromosome hampering RNA biosynthesis from some genes via competition with RNA polymerase or, vice versa, competing with inhibitors to activate transcription.
Conflict of interest statement
Figures



Similar articles
-
The DNA-Binding Protein from Starved Cells (Dps) Utilizes Dual Functions To Defend Cells against Multiple Stresses.J Bacteriol. 2015 Oct;197(19):3206-15. doi: 10.1128/JB.00475-15. Epub 2015 Jul 27. J Bacteriol. 2015. PMID: 26216848 Free PMC article.
-
Modes of Escherichia coli Dps Interaction with DNA as Revealed by Atomic Force Microscopy.PLoS One. 2015 May 15;10(5):e0126504. doi: 10.1371/journal.pone.0126504. eCollection 2015. PLoS One. 2015. PMID: 25978038 Free PMC article.
-
Regulation of Bacterial DNA Packaging in Early Stationary Phase by Competitive DNA Binding of Dps and IHF.Sci Rep. 2015 Dec 14;5:18146. doi: 10.1038/srep18146. Sci Rep. 2015. PMID: 26657062 Free PMC article.
-
Structure and function of bacterial H-NS protein.Biochem Soc Trans. 2016 Dec 15;44(6):1561-1569. doi: 10.1042/BST20160190. Biochem Soc Trans. 2016. PMID: 27913665 Review.
-
[The bacterial nucleoid].Rev Latinoam Microbiol. 1995 Jul-Sep;37(3):281-90. Rev Latinoam Microbiol. 1995. PMID: 8850347 Review. Spanish.
Cited by
-
Mass spectrometry and machine learning for the accurate diagnosis of benzylpenicillin and multidrug resistance of Staphylococcus aureus in bovine mastitis.PLoS Comput Biol. 2021 Jun 11;17(6):e1009108. doi: 10.1371/journal.pcbi.1009108. eCollection 2021 Jun. PLoS Comput Biol. 2021. PMID: 34115749 Free PMC article.
-
Network Rewiring: Physiological Consequences of Reciprocally Exchanging the Physical Locations and Growth-Phase-Dependent Expression Patterns of the Salmonella fis and dps Genes.mBio. 2020 Sep 8;11(5):e02128-20. doi: 10.1128/mBio.02128-20. mBio. 2020. PMID: 32900812 Free PMC article.
-
DNA as a Double-Coding Device for Information Conversion and Organization of a Self-Referential Unity.DNA (Basel). 2024 Nov 19;4(4):473-493. doi: 10.3390/dna4040032. DNA (Basel). 2024. PMID: 40098770 Free PMC article.
-
Live to fight another day: The bacterial nucleoid under stress.Mol Microbiol. 2025 Feb;123(2):168-175. doi: 10.1111/mmi.15272. Epub 2024 May 1. Mol Microbiol. 2025. PMID: 38690745 Review.
-
Visualization of Nucleic Acids in Microand Nanometer-Scale Biological Objects Using Analytical Electron Microscopy.Acta Naturae. 2024 Oct-Dec;16(4):38-47. doi: 10.32607/actanaturae.27483. Acta Naturae. 2024. PMID: 39877006 Free PMC article.
References
-
- Almirón M, Link AJ, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6: 2646–2654. Available: http://www.ncbi.nlm.nih.gov/pubmed/1340475. - PubMed
-
- Talukder AA, Ishihama A. Dps is a stationary phase—specific protein of Escherichia coli nucleoid. Adv Microbiol. 2014;4: 1095–1104. doi: doi: 10.4236/aim.2014.415120 - DOI
-
- Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J Bacteriol. 1999;181: 6361–6370. Available: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC103771. - PMC - PubMed
-
- Ishihama A, Kori A, Koshio E, Yamada K, Maeda H, Shimada T, et al. Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J Bacteriol. 2014;196: 2718–2727. doi: 10.1128/JB.01579-14 - DOI - PMC - PubMed
-
- Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5: 294–303. doi: 10.1038/nsb0498-294 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

