RNA-sequencing-based transcriptome and biochemical analyses of steroidal saponin pathway in a complete set of Allium fistulosum-A. cepa monosomic addition lines
- PMID: 28800607
- PMCID: PMC5553718
- DOI: 10.1371/journal.pone.0181784
RNA-sequencing-based transcriptome and biochemical analyses of steroidal saponin pathway in a complete set of Allium fistulosum-A. cepa monosomic addition lines
Erratum in
-
Correction: RNA-sequencing-based transcriptome and biochemical analyses of steroidal saponin pathway in a complete set of Allium fistulosum-A. cepa monosomic addition lines.PLoS One. 2018 Jan 2;13(1):e0190813. doi: 10.1371/journal.pone.0190813. eCollection 2018. PLoS One. 2018. PMID: 29293648 Free PMC article.
Abstract
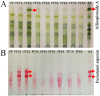
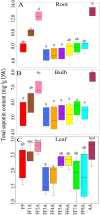
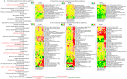
The genus Allium is a rich source of steroidal saponins, and its medicinal properties have been attributed to these bioactive compounds. The saponin compounds with diverse structures play a pivotal role in Allium's defense mechanism. Despite numerous studies on the occurrence and chemical structure of steroidal saponins, their biosynthetic pathway in Allium species is poorly understood. The monosomic addition lines (MALs) of the Japanese bunching onion (A. fistulosum, FF) with an extra chromosome from the shallot (A. cepa Aggregatum group, AA) are powerful genetic resources that enable us to understand many physiological traits of Allium. In the present study, we were able to isolate and identify Alliospiroside A saponin compound in A. fistulosum with extra chromosome 2A from shallot (FF2A) and its role in the defense mechanism against Fusarium pathogens. Furthermore, to gain molecular insight into the Allium saponin biosynthesis pathway, high-throughput RNA-Seq of the root, bulb, and leaf of AA, MALs, and FF was carried out using Illumina's HiSeq 2500 platform. An open access Allium Transcript Database (Allium TDB, http://alliumtdb.kazusa.or.jp) was generated based on RNA-Seq data. The resulting assembled transcripts were functionally annotated, revealing 50 unigenes involved in saponin biosynthesis. Differential gene expression (DGE) analyses of AA and MALs as compared with FF (as a control) revealed a strong up-regulation of the saponin downstream pathway, including cytochrome P450, glycosyltransferase, and beta-glucosidase in chromosome 2A. An understanding of the saponin compounds and biosynthesis-related genes would facilitate the development of plants with unique saponin content and, subsequently, improved disease resistance.
Conflict of interest statement
Figures


References
-
- Kamenetsky R, Rabinowitch HD. The Genus Allium: A developmental and horticultural analysis. Horticult Rev. 2006;32: 329–378.
-
- Fritsch RM, Blattner FR, Gurushidze M. New classification of Allium L. subg. Melanocrommyum (Webb & Berthel) Rouy (Alliaceae) based on molecular and morphological characters. Phyton. 2010;49: 145–220.
-
- Abdelrahman M, Hirata S, Ito S-I, Yamauchi N, Shigyo M. Compartmentation and localization of bioactive metabolites in different organs of Allium roylei. Biosci Biotechnol Biochem. 2014;7: 1112–1122. - PubMed
-
- Mostafa A, Jogaiah S, El-Sayed M, Ito S, Ikeda T, Yamauchi N, et al. Aginoside saponin, a potent antifungal compound, and secondary metabolite analyses from Allium nigrum L. Phytochem Lett. 2013;6: 274–280.
-
- Caruso G, Conti S, Villari G, Borrelli C, Melchionna G, Mintutolo M, et al. Effects of transplanting time and plant density on yield, quality and antioxidant content of onion (Allium cepa L.) in southern Italy. Sci Hortic. 2014;166: 111–120.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

