orco Mutagenesis Causes Loss of Antennal Lobe Glomeruli and Impaired Social Behavior in Ants
- PMID: 28802042
- PMCID: PMC5556950
- DOI: 10.1016/j.cell.2017.07.001
orco Mutagenesis Causes Loss of Antennal Lobe Glomeruli and Impaired Social Behavior in Ants
Abstract
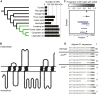
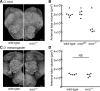
Life inside ant colonies is orchestrated with diverse pheromones, but it is not clear how ants perceive these social signals. It has been proposed that pheromone perception in ants evolved via expansions in the numbers of odorant receptors (ORs) and antennal lobe glomeruli. Here, we generate the first mutant lines in the clonal raider ant, Ooceraea biroi, by disrupting orco, a gene required for the function of all ORs. We find that orco mutants exhibit severe deficiencies in social behavior and fitness, suggesting they are unable to perceive pheromones. Surprisingly, unlike in Drosophila melanogaster, orco mutant ants also lack most of the ∼500 antennal lobe glomeruli found in wild-type ants. These results illustrate that ORs are essential for ant social organization and raise the possibility that, similar to mammals, receptor function is required for the development and/or maintenance of the highly complex olfactory processing areas in the ant brain. VIDEO ABSTRACT.
Keywords: CRISPR; Formicidae; Ooceraea biroi; chemical communication; evo-devo; mutagenesis; odorant receptors; pheromones.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Comment in
-
The MutAnts Are Here.Cell. 2017 Aug 10;170(4):601-602. doi: 10.1016/j.cell.2017.07.046. Cell. 2017. PMID: 28802035
-
Model organisms: New tools, new insights - probing social behaviour in ants.Nat Rev Genet. 2017 Oct;18(10):577. doi: 10.1038/nrg.2017.70. Epub 2017 Aug 30. Nat Rev Genet. 2017. PMID: 28852225 No abstract available.
-
CRISPR gene-editing creates wave of exotic model organisms.Nature. 2019 Apr;568(7753):441-442. doi: 10.1038/d41586-019-01300-9. Nature. 2019. PMID: 31015699 No abstract available.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Barron J, Fleet D, Beauchemin S. Performance of optical flow techniques. Int J Comput Vis. 1994;12:43–77.
-
- Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed-effects models using Eigen and S4. 2014. (R package version 1.7).
-
- Bell WJ, Parsons C, Martinko EA. Cockroach aggregation pheromones: analysis of aggregation tendency and species specificity (Orthoptera: Blattidae) J Kansas Entomol Soc. 1972;45:414–421.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

