Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses
- PMID: 28802045
- PMCID: PMC5570545
- DOI: 10.1016/j.cell.2017.07.043
Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses
Abstract
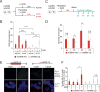
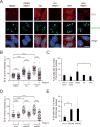
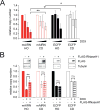
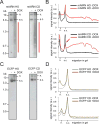
Conflicts between transcription and replication are a potent source of DNA damage. Co-transcriptional R-loops could aggravate such conflicts by creating an additional barrier to replication fork progression. Here, we use a defined episomal system to investigate how conflict orientation and R-loop formation influence genome stability in human cells. R-loops, but not normal transcription complexes, induce DNA breaks and orientation-specific DNA damage responses during conflicts with replication forks. Unexpectedly, the replisome acts as an orientation-dependent regulator of R-loop levels, reducing R-loops in the co-directional (CD) orientation but promoting their formation in the head-on (HO) orientation. Replication stress and deregulated origin firing increase the number of HO collisions leading to genome-destabilizing R-loops. Our findings connect DNA replication to R-loop homeostasis and suggest a mechanistic basis for genome instability resulting from deregulated DNA replication, observed in cancer and other disease states.
Keywords: DNA replication; DNA-damage response; R-loops; genome instability; replication stress; transcription-replication conflicts.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Transcription-Replication Conflicts: Orientation Matters.Cell. 2017 Aug 10;170(4):603-604. doi: 10.1016/j.cell.2017.07.040. Cell. 2017. PMID: 28802036
References
-
- Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. - PubMed
-
- Andersson R, Refsing Andersen P, Valen E, Core LJ, Bornholdt J, Boyd M, Heick Jensen T, Sandelin A. Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat Commun. 2014;5:5336. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

