Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis
- PMID: 28802046
- PMCID: PMC5630229
- DOI: 10.1016/j.cell.2017.07.044
Replication-Transcription Conflicts Generate R-Loops that Orchestrate Bacterial Stress Survival and Pathogenesis
Abstract
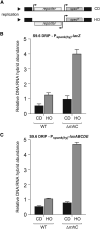
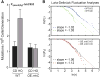
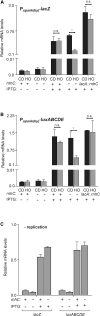
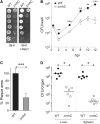
Replication-transcription collisions shape genomes, influence evolution, and promote genetic diseases. Although unclear why, head-on transcription (lagging strand genes) is especially disruptive to replication and promotes genomic instability. Here, we find that head-on collisions promote R-loop formation in Bacillus subtilis. We show that pervasive R-loop formation at head-on collision regions completely blocks replication, elevates mutagenesis, and inhibits gene expression. Accordingly, the activity of the R-loop processing enzyme RNase HIII at collision regions is crucial for stress survival in B. subtilis, as many stress response genes are head-on to replication. Remarkably, without RNase HIII, the ability of the intracellular pathogen Listeria monocytogenes to infect and replicate in hosts is weakened significantly, most likely because many virulence genes are head-on to replication. We conclude that the detrimental effects of head-on collisions stem primarily from excessive R-loop formation and that the resolution of these structures is critical for bacterial stress survival and pathogenesis.
Keywords: DNA replication; Listeria; R-loops; RNase H; accelerated evolution; gene orientation; pathogenesis; replication restart; replication-transcription conflicts; stress response.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Transcription-Replication Conflicts: Orientation Matters.Cell. 2017 Aug 10;170(4):603-604. doi: 10.1016/j.cell.2017.07.040. Cell. 2017. PMID: 28802036
References
-
- Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012;46:115–124. - PubMed
-
- Bigot A, Raynaud C, Dubail I, Dupuis M, Hossain H, Hain T, Chakraborty T, Charbit A. lmo1273, a novel gene involved in Listeria monocytogenes virulence. Microbiology. 2009;155:891–902. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

