Horizontal Dissemination of Antimicrobial Resistance Determinants in Multiple Salmonella Serotypes following Isolation from the Commercial Swine Operation Environment after Manure Application
- PMID: 28802274
- PMCID: PMC5626988
- DOI: 10.1128/AEM.01503-17
Horizontal Dissemination of Antimicrobial Resistance Determinants in Multiple Salmonella Serotypes following Isolation from the Commercial Swine Operation Environment after Manure Application
Abstract
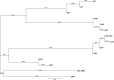
The aim of this study was to characterize the plasmids carrying antimicrobial resistance (AMR) determinants in multiple Salmonella serotypes recovered from the commercial swine farm environment after manure application on land. Manure and soil samples were collected on day 0 before and after manure application on six farms in North Carolina, and sequential soil samples were recollected on days 7, 14, and 21 from the same plots. All environmental samples were processed for Salmonella, and their plasmid contents were further characterized. A total of 14 isolates including Salmonella enterica serotypes Johannesburg (n = 2), Ohio (n = 2), Rissen (n = 1), Typhimurium var5- (n = 5), Worthington (n = 3), and 4,12:i:- (n = 1), representing different farms, were selected for plasmid analysis. Antimicrobial susceptibility testing was done by broth microdilution against a panel of 14 antimicrobials on the 14 confirmed transconjugants after conjugation assays. The plasmids were isolated by modified alkaline lysis, and PCRs were performed on purified plasmid DNA to identify the AMR determinants and the plasmid replicon types. The plasmids were sequenced for further analysis and to compare profiles and create phylogenetic trees. A class 1 integron with an ANT(2″)-Ia-aadA2 cassette was detected in the 50-kb IncN plasmids identified in S Worthington isolates. We identified 100-kb and 90-kb IncI1 plasmids in S Johannesburg and S Rissen isolates carrying the blaCMY-2 and tet(A) genes, respectively. An identical 95-kb IncF plasmid was widely disseminated among the different serotypes and across different farms. Our study provides evidence on the importance of horizontal dissemination of resistance determinants through plasmids of multiple Salmonella serotypes distributed across commercial swine farms after manure application.IMPORTANCE The horizontal gene transfer of antimicrobial resistance (AMR) determinants located on plasmids is considered to be the main reason for the rapid proliferation and spread of drug resistance. The deposition of manure generated in swine production systems into the environment is identified as a potential source of AMR dissemination. In this study, AMR gene-carrying plasmids were detected in multiple Salmonella serotypes across different commercial swine farms in North Carolina. The plasmid profiles were characterized based on Salmonella serotype donors and incompatibility (Inc) groups. We found that different Inc plasmids showed evidence of AMR gene transfer in multiple Salmonella serotypes. We detected an identical 95-kb plasmid that was widely distributed across swine farms in North Carolina. These conjugable resistance plasmids were able to persist on land after swine manure application. Our study provides strong evidence of AMR determinant dissemination present in plasmids of multiple Salmonella serotypes in the environment after manure application.
Keywords: Salmonella; antimicrobial resistance; environment; environmental microbiology; horizontal gene transfer; plasmid; swine farm.
Copyright © 2017 American Society for Microbiology.
Figures

References
-
- World Health Organization (WHO). 2015. Global action plan on antimicrobial resistance. World Health Organization, Geneva, Switzerland.
-
- World Health Organization (WHO). 2014. Antimicrobial resistance: global report on surveillance. World Health Organization, Geneva, Switzerland.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

