Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets
- PMID: 28804123
- PMCID: PMC5843901
- DOI: 10.1038/leu.2017.251
Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets
Abstract
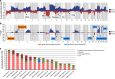
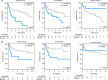
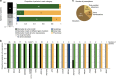
Genome studies of diffuse large B-cell lymphoma (DLBCL) have revealed a large number of somatic mutations and structural alterations. However, the clinical significance of these alterations is still not well defined. In this study, we have integrated the analysis of targeted next-generation sequencing of 106 genes and genomic copy number alterations (CNA) in 150 DLBCL. The clinically significant findings were validated in an independent cohort of 111 patients. Germinal center B-cell and activated B-cell DLBCL had a differential profile of mutations, altered pathogenic pathways and CNA. Mutations in genes of the NOTCH pathway and tumor suppressor genes (TP53/CDKN2A), but not individual genes, conferred an unfavorable prognosis, confirmed in the independent validation cohort. A gene expression profiling analysis showed that tumors with NOTCH pathway mutations had a significant modulation of downstream target genes, emphasizing the relevance of this pathway in DLBCL. An in silico drug discovery analysis recognized 69 (46%) cases carrying at least one genomic alteration considered a potential target of drug response according to early clinical trials or preclinical assays in DLBCL or other lymphomas. In conclusion, this study identifies relevant pathways and mutated genes in DLBCL and recognizes potential targets for new intervention strategies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Swerdlow S, Campo E, Harris N, Jaffe E, Pileri S, Stein H et al. World Health Organization Classification of Tumours: Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues, Fourth edition. IARC Press: Lyon, 2008.
-
- Coiffier B. Rituximab therapy in malignant lymphoma. Oncogene 2007; 26: 3603–3613. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–511. - PubMed
-
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. New Engl J Med 2002; 346: 1937–1947. - PubMed
-
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004; 103: 275–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

