Performance of objective functions and optimisation procedures for parameter estimation in system biology models
- PMID: 28804640
- PMCID: PMC5548920
- DOI: 10.1038/s41540-017-0023-2
Performance of objective functions and optimisation procedures for parameter estimation in system biology models
Abstract
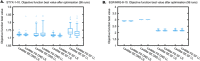
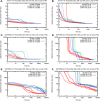
Mathematical modelling of signalling pathways aids experimental investigation in system and synthetic biology. Ever increasing data availability prompts the development of large dynamic models with numerous parameters. In this paper, we investigate how the number of unknown parameters affects the convergence of three frequently used optimisation algorithms and four objective functions. We compare objective functions that use data-driven normalisation of the simulations with those that use scaling factors. The data-driven normalisation of the simulation approach implies that simulations are normalised in the same way as the data, making both directly comparable. The scaling factor approach, which is commonly used for parameter estimation in dynamic systems, introduces scaling factors that multiply the simulations to convert them to the scale of the data. Here we show that the scaling factor approach increases, compared to data-driven normalisation of the simulations, the degree of practical non-identifiability, defined as the number of directions in the parameter space, along which parameters are not identifiable. Further, the results indicate that data-driven normalisation of the simulations greatly improve the speed of convergence of all tested algorithms when the overall number of unknown parameters is relatively large (74 parameters in our test problems). Data-driven normalisation of the simulations also markedly improve the performance of the non-gradient-based algorithm tested even when the number of unknown parameters is relatively small (10 parameters in our test problems). As the models and the unknown parameters increase in size, the data-driven normalisation of the simulation approach can be the preferred option, because it does not aggravate non-identifiability and allows for obtaining parameter estimates in a reasonable amount of time.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

