Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4
- PMID: 28805820
- PMCID: PMC5625299
- DOI: 10.1038/nm.4378
Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4
Abstract
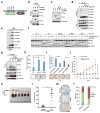
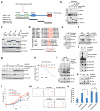
The bromodomain and extraterminal (BET) family of proteins comprises four members-BRD2, BRD3, BRD4 and the testis-specific isoform BRDT-that largely function as transcriptional coactivators and play critical roles in various cellular processes, including the cell cycle, apoptosis, migration and invasion. BET proteins enhance the oncogenic functions of major cancer drivers by elevating the expression of these drivers, such as c-Myc in leukemia, or by promoting the transcriptional activities of oncogenic factors, such as AR and ERG in prostate cancer. Pathologically, BET proteins are frequently overexpressed and are clinically linked to various types of human cancer; they are therefore being pursued as attractive therapeutic targets for selective inhibition in patients with cancer. To this end, a number of bromodomain inhibitors, including JQ1 and I-BET, have been developed and have shown promising outcomes in early clinical trials. Although resistance to BET inhibitors has been documented in preclinical models, the molecular mechanisms underlying acquired resistance are largely unknown. Here we report that cullin-3SPOP earmarks BET proteins, including BRD2, BRD3 and BRD4, for ubiquitination-mediated degradation. Pathologically, prostate cancer-associated SPOP mutants fail to interact with and promote the degradation of BET proteins, leading to their elevated abundance in SPOP-mutant prostate cancer. As a result, prostate cancer cell lines and organoids derived from individuals harboring SPOP mutations are more resistant to BET-inhibitor-induced cell growth arrest and apoptosis. Therefore, our results elucidate the tumor-suppressor role of SPOP in prostate cancer in which it acts as a negative regulator of BET protein stability and also provide a molecular mechanism for resistance to BET inhibitors in individuals with prostate cancer bearing SPOP mutations.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Comment in
-
Prostate cancer: Place your BETs.Nat Rev Urol. 2017 Oct;14(10):585. doi: 10.1038/nrurol.2017.147. Epub 2017 Aug 31. Nat Rev Urol. 2017. PMID: 28858337 No abstract available.
-
Prostate cancer: BET inhibitors - SPOP right there!Nat Rev Cancer. 2017 Oct;17(10):574-575. doi: 10.1038/nrc.2017.80. Epub 2017 Sep 8. Nat Rev Cancer. 2017. PMID: 28883513 No abstract available.
-
Re: Prostate Cancer-Associated SPOP Mutations Confer Resistance to BET Inhibitors through Stabilization of BRD4.J Urol. 2018 Apr;199(4):895-896. doi: 10.1016/j.juro.2018.01.003. Epub 2018 Jan 9. J Urol. 2018. PMID: 29642362 No abstract available.
References
-
- Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. - PubMed
-
- Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. - PubMed
-
- Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586:2692–2704. - PubMed
-
- Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. - PubMed
MeSH terms
Substances
Grants and funding
- R01 CA200651/CA/NCI NIH HHS/United States
- R00 CA181342/CA/NCI NIH HHS/United States
- R01 CA193837/CA/NCI NIH HHS/United States
- R01 GM094777/GM/NIGMS NIH HHS/United States
- R01 CA200573/CA/NCI NIH HHS/United States
- P50 CA092629/CA/NCI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- K08 CA140946/CA/NCI NIH HHS/United States
- R01 CA208100/CA/NCI NIH HHS/United States
- K99 CA207867/CA/NCI NIH HHS/United States
- R01 CA177910/CA/NCI NIH HHS/United States
- R01 GM089763/GM/NIGMS NIH HHS/United States
- U54 CA224079/CA/NCI NIH HHS/United States
- P50 CA069568/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

