Parvovirus B19 integration into human CD36+ erythroid progenitor cells
- PMID: 28806616
- PMCID: PMC5623651
- DOI: 10.1016/j.virol.2017.08.011
Parvovirus B19 integration into human CD36+ erythroid progenitor cells
Abstract
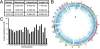
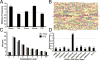
The pathogenic autonomous human parvovirus B19 (B19V) productively infects erythroid progenitor cells (EPCs). Functional similarities between B19V nonstructural protein (NS1), a DNA binding endonuclease, and the Rep proteins of Adeno-Associated Virus (AAV) led us to hypothesize that NS1 may facilitate targeted nicking of the human genome and B19 vDNA integration. We adapted an integration capture sequencing protocol (IC-Seq) to screen B19V infected human CD36+ EPCs for viral integrants, and discovered 40,000 unique B19V integration events distributed throughout the human genome. Computational analysis of integration patterns revealed strong correlations with gene intronic regions, H3K9me3 sites, and the identification of 41 base pair consensus sequence with an octanucleotide core motif. The octanucleotide core has homology to a single region of B19V, adjacent to the P6 promoter TATA box. We present the first direct evidence that B19V infection of erythroid progenitor cells disrupts the human genome and facilitates viral DNA integration.
Keywords: B19; High throughput sequencing; Human erythroid progenitor cell; Integration; Latency; Parvovirus.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


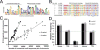
References
-
- Adamson-Small LA, Ignatovich IV, Laemmerhirt MG, Hobbs JA. Persistent parvovirus B19 infection in non-erythroid tissues: possible role in the inflammatory and disease process. Virus Res. 2014;190:8–16. - PubMed
-
- Bonvicini F, Filippone C, Manaresi E, Zerbini M, Musiani M, Gallinella G. HepG2 hepatocellular carcinoma cells are a non-permissive system for B19 virus infection. J Gen Virol. 2008;89:3034–3038. - PubMed
-
- Brown KE, Anderson SM, Young NS. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262:114–117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

