Expansion and evolutionary patterns of cysteine-rich peptides in plants
- PMID: 28806914
- PMCID: PMC5557327
- DOI: 10.1186/s12864-017-3948-3
Expansion and evolutionary patterns of cysteine-rich peptides in plants
Abstract
Background: Cysteine-rich peptides (CRPs) are gaining recognition as regulators of cell-cell communication in plants.
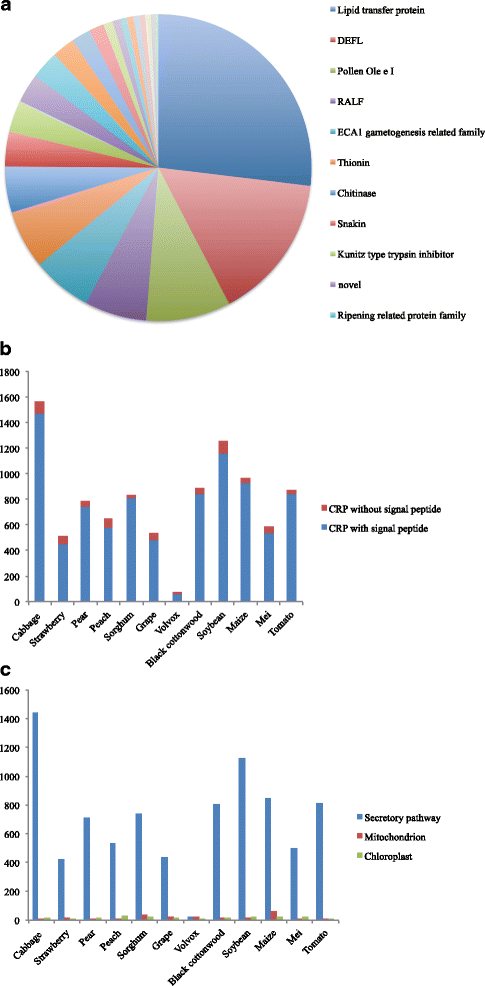
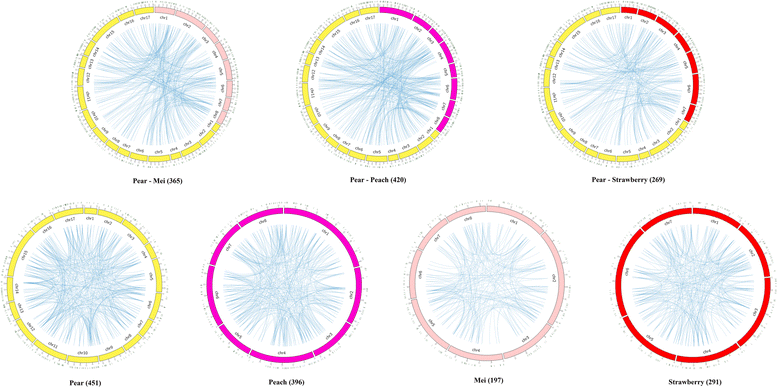
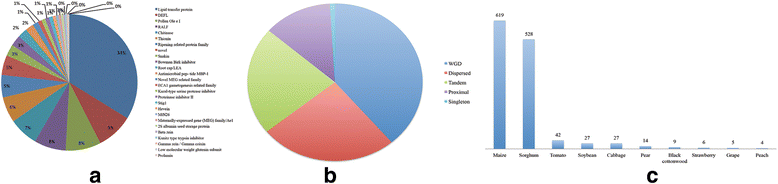
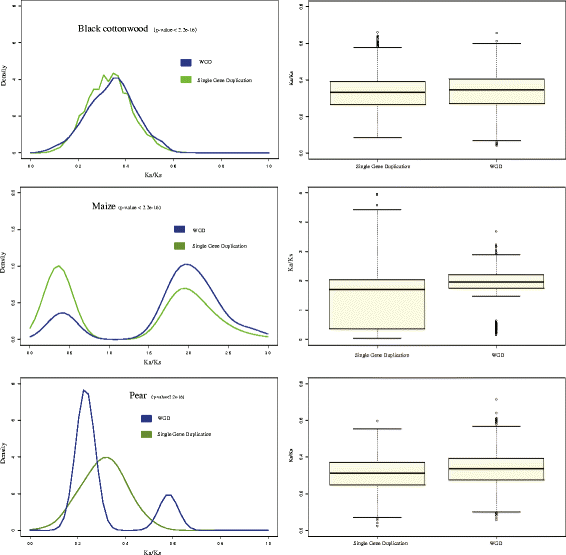
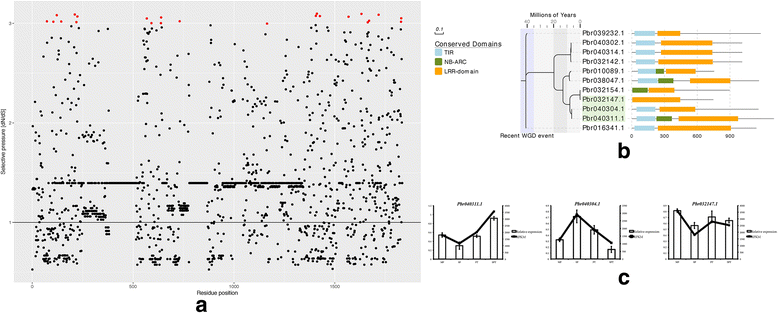
Results: We identified 9556 CRPs in 12 plant species and analysed their evolutionary patterns. In most angiosperm plants, whole genome duplication and segmental duplication are the major factors driving the expansion of CRP family member genes, especially signal peptides. About 30% of the CRP genes were found clustered on the chromosomes, except in maize (Zea mays). Considerable collinearities between CRP genes between or within species reveal several syntenic regions on the chromosomes. Different subfamilies display diverse evolutionary rates, suggesting that these subfamilies are subjected to different selective pressures. CRPs in different duplication models also show contrasting evolutionary rates, although the underlying mechanism is unclear because of the complexity of gene evolution. The 1281 positively selected genes identified are probably generated within a certain period of time. While most of these belonged to maize and sorghum (Sorghum bicolor), new CRP functions would also be expected. Up-regulation of 10 CRPs was observed in self-pollinated pear pistils and pollen tubes under self S-RNase treatments in vitro. The expression divergence between different CRP gene duplication types suggests that different duplication mechanisms affected the fate of the duplicated CRPs.
Conclusion: Our analyses of the evolution of the CRP gene family provides a unique view of the evolution of this large gene family.
Keywords: Clustered genes; Cysteine-rich peptide; Divergent evolution pattern; Expression divergence; Gene duplication; Positive selection; Self-incompatibility.
Conflict of interest statement
Ethics approval and consent to participate
The genome database of Chinese white pear is acquired from Center of Pear Engineering Technology Research; the pollen of ‘Dangshansuli’ and ‘Fengshui’ is collected from Jiangpu farm in Nanjing agricultural university. The genome database and pollen are applied for our research under the permission of Center of Pear Engineering Technology Research.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

