A genome Tree of Life for the Fungi kingdom
- PMID: 28808018
- PMCID: PMC5584464
- DOI: 10.1073/pnas.1711939114
A genome Tree of Life for the Fungi kingdom
Abstract
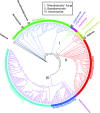
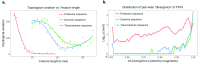
Fungi belong to one of the largest and most diverse kingdoms of living organisms. The evolutionary kinship within a fungal population has so far been inferred mostly from the gene-information-based trees ("gene trees"), constructed commonly based on the degree of differences of proteins or DNA sequences of a small number of highly conserved genes common among the population by a multiple sequence alignment (MSA) method. Since each gene evolves under different evolutionary pressure and time scale, it has been known that one gene tree for a population may differ from other gene trees for the same population depending on the subjective selection of the genes. Within the last decade, a large number of whole-genome sequences of fungi have become publicly available, which represent, at present, the most fundamental and complete information about each fungal organism. This presents an opportunity to infer kinship among fungi using a whole-genome information-based tree ("genome tree"). The method we used allows comparison of whole-genome information without MSA, and is a variation of a computational algorithm developed to find semantic similarities or plagiarism in two books, where we represent whole-genomic information of an organism as a book of words without spaces. The genome tree reveals several significant and notable differences from the gene trees, and these differences invoke new discussions about alternative narratives for the evolution of some of the currently accepted fungal groups.
Keywords: alignment-free method; divergence tree; feature frequency profile; fungal phylogeny; proteome tree.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Taylor JW, et al. The fungi. In: J Cracraft, MJ Donoghue., editors. Assembling the Tree of Life. Oxford Univ Press; New York: 2004.
-
- Petersen JH. The Kingdom of Fungi. Princeton Univ Press; Princeton, NJ: 2013.
-
- James TY, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. - PubMed
-
- Hibbett DS, et al. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

