A bioarchitectonic approach to the modular engineering of metabolism
- PMID: 28808103
- PMCID: PMC5566884
- DOI: 10.1098/rstb.2016.0387
A bioarchitectonic approach to the modular engineering of metabolism
Abstract
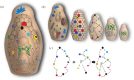
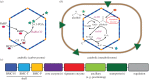
Dissociating the complexity of metabolic processes into modules is a shift in focus from the single gene/gene product to functional and evolutionary units spanning the scale of biological organization. When viewing the levels of biological organization through this conceptual lens, modules are found across the continuum: domains within proteins, co-regulated groups of functionally associated genes, operons, metabolic pathways and (sub)cellular compartments. Combining modules as components or subsystems of a larger system typically leads to increased complexity and the emergence of new functions. By virtue of their potential for 'plug and play' into new contexts, modules can be viewed as units of both evolution and engineering. Through consideration of lessons learned from recent efforts to install new metabolic modules into cells and the emerging understanding of the structure, function and assembly of protein-based organelles, bacterial microcompartments, a structural bioengineering approach is described: one that builds from an architectural vocabulary of protein domains. This bioarchitectonic approach to engineering cellular metabolism can be applied to microbial cell factories, used in the programming of members of synthetic microbial communities or used to attain additional levels of metabolic organization in eukaryotic cells for increasing primary productivity and as the foundation of a green economy.This article is part of the themed issue 'Enhancing photosynthesis in crop plants: targets for improvement'.
Keywords: bacterial microcompartment; carboxysome; synthetic biology.
© 2017 The Author(s).
Conflict of interest statement
I declare I have no competing interests.
Figures

Similar articles
-
Carboxysomes: metabolic modules for CO2 fixation.FEMS Microbiol Lett. 2017 Oct 2;364(18). doi: 10.1093/femsle/fnx176. FEMS Microbiol Lett. 2017. PMID: 28934381 Review.
-
Bacterial microcompartments and the modular construction of microbial metabolism.Trends Microbiol. 2015 Jan;23(1):22-34. doi: 10.1016/j.tim.2014.10.003. Epub 2014 Nov 14. Trends Microbiol. 2015. PMID: 25455419 Review.
-
Interrelated modules in cyanobacterial photosynthesis: the carbon-concentrating mechanism, photorespiration, and light perception.J Exp Bot. 2016 May;67(10):2931-40. doi: 10.1093/jxb/erw162. Epub 2016 Apr 25. J Exp Bot. 2016. PMID: 27117337
-
Bacterial microcompartments as metabolic modules for plant synthetic biology.Plant J. 2016 Jul;87(1):66-75. doi: 10.1111/tpj.13166. Epub 2016 Jun 20. Plant J. 2016. PMID: 26991644
-
Current Status of Microbial Phenylethanoid Biosynthesis.J Microbiol Biotechnol. 2018 Aug 28;28(8):1225-1232. doi: 10.4014/jmb.1805.05021. J Microbiol Biotechnol. 2018. PMID: 29996621 Review.
Cited by
-
Bacterial microcompartments.Nat Rev Microbiol. 2018 May;16(5):277-290. doi: 10.1038/nrmicro.2018.10. Epub 2018 Mar 5. Nat Rev Microbiol. 2018. PMID: 29503457 Free PMC article. Review.
-
Engineering spatiotemporal organization and dynamics in synthetic cells.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2021 May;13(3):e1685. doi: 10.1002/wnan.1685. Epub 2020 Nov 21. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2021. PMID: 33219745 Free PMC article. Review.
-
Fixing carbon: To alleviate climate change, scientists are exploring ways to harness nature's ability to capture CO2 from the atmosphere.EMBO Rep. 2019 Feb;20(2):e47580. doi: 10.15252/embr.201847580. Epub 2019 Jan 3. EMBO Rep. 2019. PMID: 30606749 Free PMC article. Review.
-
Bacterial microcompartments: catalysis-enhancing metabolic modules for next generation metabolic and biomedical engineering.BMC Biol. 2019 Oct 10;17(1):79. doi: 10.1186/s12915-019-0691-z. BMC Biol. 2019. PMID: 31601225 Free PMC article. Review.
-
Spatially organizing biochemistry: choosing a strategy to translate synthetic biology to the factory.Sci Rep. 2018 May 29;8(1):8196. doi: 10.1038/s41598-018-26399-0. Sci Rep. 2018. PMID: 29844460 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

