A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9
- PMID: 28808686
- PMCID: PMC5547770
- DOI: 10.1126/sciadv.aao0027
A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9
Abstract
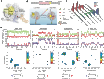
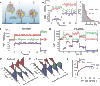
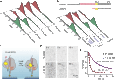
The Cas9 endonuclease is widely used for genome engineering applications by programming its single-guide RNA, and ongoing work is aimed at improving the accuracy and efficiency of DNA targeting. DNA cleavage of Cas9 is controlled by the conformational state of the HNH nuclease domain, but the mechanism that governs HNH activation at on-target DNA while reducing cleavage activity at off-target sites remains poorly understood. Using single-molecule Förster resonance energy transfer, we identified an intermediate state of Streptococcus pyogenes Cas9, representing a conformational checkpoint between DNA binding and cleavage. Upon DNA binding, the HNH domain transitions between multiple conformations before docking into its active state. HNH docking requires divalent cations, but not strand scission, and this docked conformation persists following DNA cleavage. Sequence mismatches between the DNA target and guide RNA prevent transitions from the checkpoint intermediate to the active conformation, providing selective avoidance of DNA cleavage at stably bound off-target sites.
Figures

References
-
- Mohanraju P., Makarova K. S., Zetsche B., Zhang F., Koonin E. V., van der Oost J., Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147 (2016). - PubMed
-
- Wright A. V., Nuñez J. K., Doudna J. A., Biology and Applications of CRISPR Systems: Harnessing nature’s toolbox for genome engineering. Cell 164, 29–44 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

