Glucose Limitation Alters Glutamine Metabolism in MUC1-Overexpressing Pancreatic Cancer Cells
- PMID: 28809118
- PMCID: PMC5634392
- DOI: 10.1021/acs.jproteome.7b00246
Glucose Limitation Alters Glutamine Metabolism in MUC1-Overexpressing Pancreatic Cancer Cells
Abstract
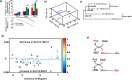
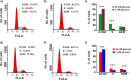
Pancreatic cancer cells overexpressing Mucin 1 (MUC1) rely on aerobic glycolysis and, correspondingly, are dependent on glucose for survival. Our NMR metabolomics comparative analysis of control (S2-013.Neo) and MUC1-overexpressing (S2-013.MUC1) cells demonstrates that MUC1 reprograms glutamine metabolism upon glucose limitation. The observed alteration in glutamine metabolism under glucose limitation was accompanied by a relative decrease in the proliferation of MUC1-overexpressing cells compared with steady-state conditions. Moreover, glucose limitation induces G1 phase arrest where S2-013.MUC1 cells fail to enter S phase and synthesize DNA because of a significant disruption in pyrimidine nucleotide biosynthesis. Our metabolomics analysis indicates that glutamine is the major source of oxaloacetate in S2-013.Neo and S2-013.MUC1 cells, where oxaloacetate is converted to aspartate, an important metabolite for pyrimidine nucleotide biosynthesis. However, glucose limitation impedes the flow of glutamine carbons into the pyrimidine nucleotide rings and instead leads to a significant accumulation of glutamine-derived aspartate in S2-013.MUC1 cells.
Keywords: MUC1 overexpression; NMR metabolomics; cancer metabolism; glucose limitation; glutamine metabolism.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Singh P. K.; Behrens M. E.; Eggers J. P.; Cerny R. L.; Bailey J. M.; Shanmugam K.; Gendler S. J.; Bennett E. P.; Hollingsworth M. A. Phosphorylation of MUC1 by Met modulates interaction with p53 and MMP1 expression. J. Biol. Chem. 2008, 283 (40), 26985–26995. 10.1074/jbc.M805036200. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

