Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis
- PMID: 28809393
- PMCID: PMC5587856
- DOI: 10.1038/cr.2017.99
Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis
Abstract
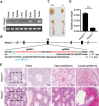
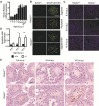
N6-methyladenosine (m6A) is the most common internal modification in eukaryotic mRNA. It is dynamically installed and removed, and acts as a new layer of mRNA metabolism, regulating biological processes including stem cell pluripotency, cell differentiation, and energy homeostasis. m6A is recognized by selective binding proteins; YTHDF1 and YTHDF3 work in concert to affect the translation of m6A-containing mRNAs, YTHDF2 expedites mRNA decay, and YTHDC1 affects the nuclear processing of its targets. The biological function of YTHDC2, the final member of the YTH protein family, remains unknown. We report that YTHDC2 selectively binds m6A at its consensus motif. YTHDC2 enhances the translation efficiency of its targets and also decreases their mRNA abundance. Ythdc2 knockout mice are infertile; males have significantly smaller testes and females have significantly smaller ovaries compared to those of littermates. The germ cells of Ythdc2 knockout mice do not develop past the zygotene stage and accordingly, Ythdc2 is upregulated in the testes as meiosis begins. Thus, YTHDC2 is an m6A-binding protein that plays critical roles during spermatogenesis.
Figures


References
-
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science 2005; 309:1514–1518. - PubMed
-
- Wei C, Moss B. Nucleotide Sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry 1977; 16:1672–1676. - PubMed
-
- Wei C, Gershowitz A, Moss B. 5′-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 1976; 15:397–401. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

