Parallel Evolution of Sperm Hyper-Activation Ca2+ Channels
- PMID: 28810709
- PMCID: PMC5553355
- DOI: 10.1093/gbe/evx131
Parallel Evolution of Sperm Hyper-Activation Ca2+ Channels
Abstract
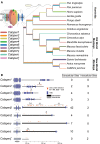
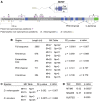
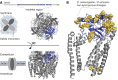
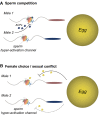
Sperm hyper-activation is a dramatic change in sperm behavior where mature sperm burst into a final sprint in the race to the egg. The mechanism of sperm hyper-activation in many metazoans, including humans, consists of a jolt of Ca2+ into the sperm flagellum via CatSper ion channels. Surprisingly, all nine CatSper genes have been independently lost in several animal lineages. In Drosophila, sperm hyper-activation is performed through the cooption of the polycystic kidney disease 2 (pkd2) Ca2+ channel. The parallels between CatSpers in primates and pkd2 in Drosophila provide a unique opportunity to examine the molecular evolution of the sperm hyper-activation machinery in two independent, nonhomologous calcium channels separated by > 500 million years of divergence. Here, we use a comprehensive phylogenomic approach to investigate the selective pressures on these sperm hyper-activation channels. First, we find that the entire CatSper complex evolves rapidly under recurrent positive selection in primates. Second, we find that pkd2 has parallel patterns of adaptive evolution in Drosophila. Third, we show that this adaptive evolution of pkd2 is driven by its role in sperm hyper-activation. These patterns of selection suggest that the evolution of the sperm hyper-activation machinery is driven by sexual conflict with antagonistic ligands that modulate channel activity. Together, our results add sperm hyper-activation channels to the class of fast evolving reproductive proteins and provide insights into the mechanisms used by the sexes to manipulate sperm behavior.
Keywords: CatSper; evolutionary arms race; parallel evolution; reproductive proteins; sexual conflict; sperm hyper-activation.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
References
-
- Cai Y, 1999. Identification and characterization of polycystin-2, the PKD2 gene product. J Biol Chem. 274(40):28557–28565. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

