Validation of β-lactam minimum inhibitory concentration predictions for pneumococcal isolates with newly encountered penicillin binding protein (PBP) sequences
- PMID: 28810827
- PMCID: PMC5558719
- DOI: 10.1186/s12864-017-4017-7
Validation of β-lactam minimum inhibitory concentration predictions for pneumococcal isolates with newly encountered penicillin binding protein (PBP) sequences
Abstract
Background: Genomic sequence-based deduction of antibiotic minimum inhibitory concentration (MIC) has great potential to enhance the speed and sensitivity of antimicrobial susceptibility testing. We previously developed a penicillin-binding protein (PBP) typing system and two methods (Random Forest (RF) and Mode MIC (MM)) that accurately predicted β-lactam MICs for pneumococcal isolates carrying a characterized PBP sequence type (phenotypic β-lactam MICs known for at least one isolate of this PBP type). This study evaluates the prediction performance for previously uncharacterized (new) PBP types and the probability of encountering new PBP types, both of which impact the overall prediction accuracy.
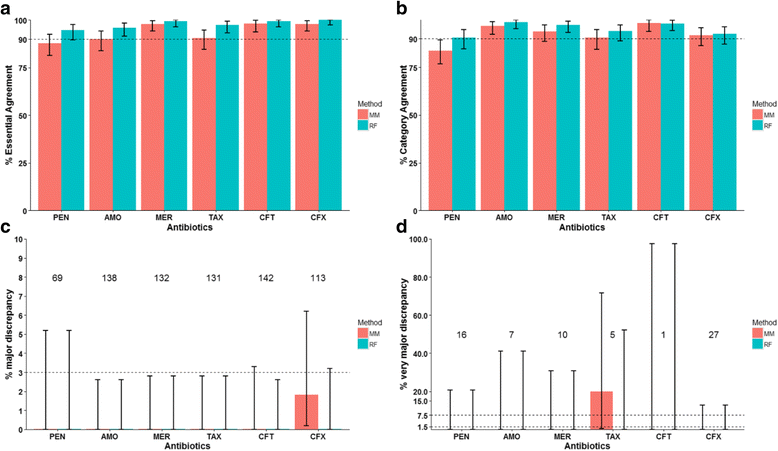
Results: The MM and RF methods were used to predict MICs of 4309 previously reported pneumococcal isolates in 2 datasets and the results were compared to the known broth microdilution MICs to 6 β-lactams. Based on a method that specifically evaluated predictions for new PBP types, the RF results were more accurate than MM results for new PBP types and showed percent essential agreement (MICs agree within ±1 dilution) >97%, percent category agreement (interpretive results agree) >93%, major discrepancy (sensitive isolate predicted as resistant) rate < 1.2%, and very major discrepancy (resistant isolate predicted as sensitive) rate < 1.4% for all 6 β-lactams. The identification of new PBP types over time was well approximated by a diminishingly increasing curve (Pearson's r = 0.99) and minimally impacted overall MIC prediction performance.
Conclusions: MIC prediction using the RF method could be an accurate alternative of phenotypic susceptibility testing even in the presence of previously uncharacterized PBP types.
Keywords: Minimum inhibitory concentration (MIC); Penicillin binding protein typing (PBP typing); Predictive modeling; Streptococcus Pneumoniae; β-lactam antibiotics.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Koser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M, Holden MT, Dougan G, Bentley SD, Parkhill J, et al. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 2012;8(8) doi: 10.1371/journal.ppat.1002824. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

