Angular reconstitution-based 3D reconstructions of nanomolecular structures from superresolution light-microscopy images
- PMID: 28811371
- PMCID: PMC5584428
- DOI: 10.1073/pnas.1704908114
Angular reconstitution-based 3D reconstructions of nanomolecular structures from superresolution light-microscopy images
Abstract
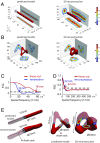
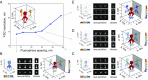
Superresolution light microscopy allows the imaging of labeled supramolecular assemblies at a resolution surpassing the classical diffraction limit. A serious limitation of the superresolution approach is sample heterogeneity and the stochastic character of the labeling procedure. To increase the reproducibility and the resolution of the superresolution results, we apply multivariate statistical analysis methods and 3D reconstruction approaches originally developed for cryogenic electron microscopy of single particles. These methods allow for the reference-free 3D reconstruction of nanomolecular structures from two-dimensional superresolution projection images. Since these 2D projection images all show the structure in high-resolution directions of the optical microscope, the resulting 3D reconstructions have the best possible isotropic resolution in all directions.
Keywords: 3D reconstruction; DNA origami; multivariate statistical analysis; structural biology; superresolution microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Schmidt C, et al. The cryo-EM structure of a ribosome-Ski2-Ski3-Ski8 helicase complex. Science. 2016;354:1431–1433. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

